PHYTOCHROME-INTERACTING FACTORs trigger environmentally responsive chromatin dynamics in plants
- PMID: 34140685
- PMCID: PMC9169284
- DOI: 10.1038/s41588-021-00882-3
PHYTOCHROME-INTERACTING FACTORs trigger environmentally responsive chromatin dynamics in plants
Erratum in
-
Author Correction: PHYTOCHROME-INTERACTING FACTORs trigger environmentally responsive chromatin dynamics in plants.Nat Genet. 2023 Feb;55(2):355. doi: 10.1038/s41588-023-01299-w. Nat Genet. 2023. PMID: 36690790 No abstract available.
Abstract
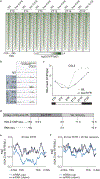
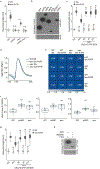
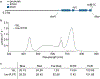
The interplay between light receptors and PHYTOCHROME-INTERACTING FACTORs (PIFs) serves as a regulatory hub that perceives and integrates environmental cues into transcriptional networks of plants1,2. Although occupancy of the histone variant H2A.Z and acetylation of histone H3 have emerged as regulators of environmentally responsive gene networks, how these epigenomic features interface with PIF activity is poorly understood3-7. By taking advantage of rapid and reversible light-mediated manipulation of PIF7 subnuclear localization and phosphorylation, we simultaneously assayed the DNA-binding properties of PIF7, as well as its impact on chromatin dynamics genome wide. We found that PIFs act rapidly to reshape the H2A.Z and H3K9ac epigenetic landscape in response to a change in light quality. Furthermore, we discovered that PIFs achieve H2A.Z removal through direct interaction with EIN6 ENHANCER (EEN), the Arabidopsis thaliana homolog of the chromatin remodeling complex subunit INO80 Subunit 6 (Ies6). Thus, we describe a PIF-INO80 regulatory module that is an intermediate step for allowing plants to change their growth trajectory in response to environmental changes.
© 2021. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures

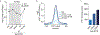




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

