Assessing the Response of Ruminal Bacterial and Fungal Microbiota to Whole-Rumen Contents Exchange in Dairy Cows
- PMID: 34140943
- PMCID: PMC8203821
- DOI: 10.3389/fmicb.2021.665776
Assessing the Response of Ruminal Bacterial and Fungal Microbiota to Whole-Rumen Contents Exchange in Dairy Cows
Abstract
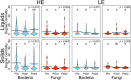
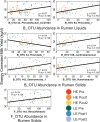
A major goal for the dairy industry is to improve overall milk production efficiency (MPE). With the advent of next-generation sequencing and advanced methods for characterizing microbial communities, efforts are underway to improve MPE by manipulating the rumen microbiome. Our previous work demonstrated that a near-total exchange of whole rumen contents between pairs of lactating Holstein dairy cows of disparate MPE resulted in a reversal of MPE status for ∼10 days: historically high-efficiency cows decreased in MPE, and historically low-efficiency cows increased in MPE. Importantly, this switch in MPE status was concomitant with a reversal in the ruminal bacterial microbiota, with the newly exchanged bacterial communities reverting to their pre-exchange state. However, this work did not include an in-depth analysis of the microbial community response or an interrogation of specific taxa correlating to production metrics. Here, we sought to better understand the response of rumen communities to this exchange protocol, including consideration of the rumen fungi. Rumen samples were collected from 8 days prior to, and 56 days following the exchange and were subjected to 16S rRNA and ITS amplicon sequencing to assess bacterial and fungal community composition, respectively. Our results show that the ruminal fungal community did not differ significantly between hosts of disparate efficiency prior to the exchange, and no change in community structure was observed over the time course. Correlation of microbial taxa to production metrics identified one fungal operational taxonomic unit (OTU) in the genus Neocallimastix that correlated positively to MPE, and several bacterial OTUs classified to the genus Prevotella. Within the Prevotella, Prevotella_1 was found to be more abundant in high-efficiency cows whereas Prevotella_7 was more abundant in low-efficiency cows. Overall, our results suggest that the rumen bacterial community is a primary microbial driver of host efficiency, that the ruminal fungi may not have as significant a role in MPE as previously thought, and that more work is needed to better understand the functional roles of specific ruminal microbial community members in modulating MPE.
Keywords: dairy cattle; milk production efficiency; rumen; ruminal contents exchange; ruminal microbiota.
Copyright © 2021 Cox, Deblois and Suen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Bray R. J., Curtis J. T. (1957). An ordination of the upland forest communities of Southern Wisconsin. Ecol. Monogr. 27 325–349.
-
- Chao A. (1984). Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 11 265–270.
LinkOut - more resources
Full Text Sources

