Genome-Wide Variants Associated With Longitudinal Survival Outcomes Among Individuals With Coronary Artery Disease
- PMID: 34140969
- PMCID: PMC8204081
- DOI: 10.3389/fgene.2021.661497
Genome-Wide Variants Associated With Longitudinal Survival Outcomes Among Individuals With Coronary Artery Disease
Erratum in
-
Corrigendum: Genome-Wide Variants Associated With Longitudinal Survival Outcomes Among Individuals With Coronary Artery Disease.Front Genet. 2021 Jul 27;12:726466. doi: 10.3389/fgene.2021.726466. eCollection 2021. Front Genet. 2021. PMID: 34386044 Free PMC article.
Abstract
Objective: Coronary artery disease (CAD) is an age-associated condition that greatly increases the risk of mortality. The purpose of this study was to identify gene variants associated with all-cause mortality among individuals with clinically phenotyped CAD using a genome-wide screening approach.
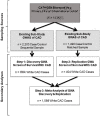
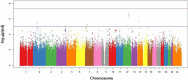
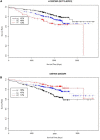
Approach and results: We performed discovery (n = 1,099), replication (n = 404), and meta-analyses (N = 1,503) for association of genomic variants with survival outcome using secondary data from White participants with CAD from two GWAS sub-studies of the Duke Catheterization Genetics Biorepository. We modeled time from catheterization to death or last follow-up (median 7.1 years, max 12 years) using Cox multivariable regression analysis. Target statistical screening thresholds were p × 10-8 for the discovery phase and Bonferroni-calculated p-values for the replication (p < 5.3 × 10-4) and meta-analysis (p < 1.6 × 10-3) phases. Genome-wide analysis of 785,945 autosomal SNPs revealed two SNPs (rs13007553 and rs587936) that had the same direction of effect across all three phases of the analysis, with suggestive p-value association in discovery and replication and significant meta-analysis association in models adjusted for clinical covariates. The rs13007553 SNP variant, LINC01250, which resides between MYTIL and EIPR1, conferred increased risk for all-cause mortality even after controlling for clinical covariates [HR 1.47, 95% CI 1.17-1.86, p(adj) = 1.07 × 10-3 (discovery), p(adj) = 0.03 (replication), p(adj) = 9.53 × 10-5 (meta-analysis)]. MYT1L is involved in neuronal differentiation. TSSC1 is involved in endosomal recycling and is implicated in breast cancer. The rs587936 variant annotated to DAB2IP was associated with increased survival time [HR 0.65, 95% CI 0.51-0.83, p(adj) = 4.79 × 10-4 (discovery), p(adj) = 0.02 (replication), p(adj) = 2.25 × 10-5 (meta-analysis)]. DAB2IP is a ras/GAP tumor suppressor gene which is highly expressed in vascular tissue. DAB2IP has multiple lines of evidence for protection against atherosclerosis.
Conclusion: Replicated findings identified two candidate genes for further study regarding association with survival in high-risk CAD patients: novel loci LINC01250 (rs13007553) and biologically relevant candidate DAB2IP (rs587936). These candidates did not overlap with validated longevity candidate genes. Future research could further define the role of common variants in survival outcomes for people with CAD and, ultimately, improve longitudinal outcomes for these patients.
Keywords: age-related disease; candidate gene analyses; coronary artery disease; genome-wide association study; survival analysis.
Copyright © 2021 Dungan, Qin, Hurdle, Haynes, Hauser and Kraus.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Aouizerat B. E., Vittinghoff E., Musone S. L., Pawlikowska L., Kwok P.-Y., Olgin J. E., et al. (2011). GWAS for discovery and replication of genetic loci associated with sudden cardiac arrest in patients with coronary artery disease. BMC Cardiovasc. Disord. 11:29. 10.1186/1471-2261-11-29 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

