Advanced tandem mass spectrometry in metabolomics and lipidomics-methods and applications
- PMID: 34142202
- PMCID: PMC8440309
- DOI: 10.1007/s00216-021-03425-1
Advanced tandem mass spectrometry in metabolomics and lipidomics-methods and applications
Abstract
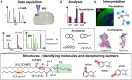
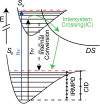
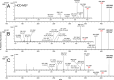
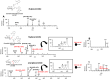
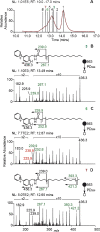
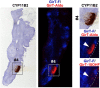
Metabolomics and lipidomics are new drivers of the omics era as molecular signatures and selected analytes allow phenotypic characterization and serve as biomarkers, respectively. The growing capabilities of untargeted and targeted workflows, which primarily rely on mass spectrometric platforms, enable extensive charting or identification of bioactive metabolites and lipids. Structural annotation of these compounds is key in order to link specific molecular entities to defined biochemical functions or phenotypes. Tandem mass spectrometry (MS), first and foremost collision-induced dissociation (CID), is the method of choice to unveil structural details of metabolites and lipids. But CID fragment ions are often not sufficient to fully characterize analytes. Therefore, recent years have seen a surge in alternative tandem MS methodologies that aim to offer full structural characterization of metabolites and lipids. In this article, principles, capabilities, drawbacks, and first applications of these "advanced tandem mass spectrometry" strategies will be critically reviewed. This includes tandem MS methods that are based on electrons, photons, and ion/molecule, as well as ion/ion reactions, combining tandem MS with concepts from optical spectroscopy and making use of derivatization strategies. In the final sections of this review, the first applications of these methodologies in combination with liquid chromatography or mass spectrometry imaging are highlighted and future perspectives for research in metabolomics and lipidomics are discussed.
Keywords: Biopolymers/lipids; HPLC; Lipidomics; Mass spectrometry imaging; Metabolomics; Tandem mass spectrometry.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Han X. Lipidomics for studying metabolism. Nat Rev Endocrinol. 2016;12:668–679. - PubMed
-
- Sperber H, Mathieu J, Wang Y, Ferreccio A, Hesson J, Xu Z, Fischer KA, Devi A, Detraux D, Gu H, Battle SL, Showalter M, Valensisi C, Bielas JH, Ericson NG, Margaretha L, Robitaille AM, Margineantu D, Fiehn O, Hockenbery D, Blau CA, Raftery D, Margolin AA, Hawkins RD, Moon RT, Ware CB, Ruohola-Baker H. The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat Cell Biol. 2015;17:1523–1535. - PMC - PubMed
-
- Nič M, Jirát J, Košata B, Jenkins A, McNaught A, editors. IUPAC compendium of chemical terminology. Research Triagle Park: IUPAC; 2009.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

