The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN
- PMID: 34142705
- PMCID: PMC8286842
- DOI: 10.1042/BCJ20210160
The structures of E. coli NfsA bound to the antibiotic nitrofurantoin; to 1,4-benzoquinone and to FMN
Abstract
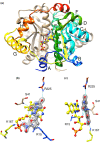
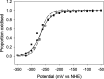
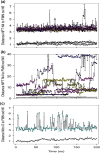
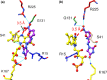
NfsA is a dimeric flavoprotein that catalyses the reduction in nitroaromatics and quinones by NADPH. This reduction is required for the activity of nitrofuran antibiotics. The crystal structure of free Escherichia coli NfsA and several homologues have been determined previously, but there is no structure of the enzyme with ligands. We present here crystal structures of oxidised E. coli NfsA in the presence of several ligands, including the antibiotic nitrofurantoin. Nitrofurantoin binds with the furan ring, rather than the nitro group that is reduced, near the N5 of the FMN. Molecular dynamics simulations show that this orientation is only favourable in the oxidised enzyme, while potentiometry suggests that little semiquinone is formed in the free protein. This suggests that the reduction occurs by direct hydride transfer from FMNH- to nitrofurantoin bound in the reverse orientation to that in the crystal structure. We present a model of nitrofurantoin bound to reduced NfsA in a viable hydride transfer orientation. The substrate 1,4-benzoquinone and the product hydroquinone are positioned close to the FMN N5 in the respective crystal structures with NfsA, suitable for reaction, but are mobile within the active site. The structure with a second FMN, bound as a ligand, shows that a mobile loop in the free protein forms a phosphate-binding pocket. NfsA is specific for NADPH and a similar conformational change, forming a phosphate-binding pocket, is likely to also occur with the natural cofactor.
Keywords: antibiotic; flavoprotein; nitroreductase; oxidation–reduction.
© 2021 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures

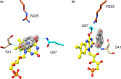

Comment in
-
Flavin oxidation state impacts on nitrofuran antibiotic binding orientation in nitroreductases.Biochem J. 2021 Sep 30;478(18):3423-3428. doi: 10.1042/BCJ20210489. Biochem J. 2021. PMID: 34554213
Similar articles
-
Oxygen-insensitive nitroreductase E. coli NfsA, but not NfsB, is inhibited by fumarate.Proteins. 2023 May;91(5):585-592. doi: 10.1002/prot.26451. Epub 2022 Dec 13. Proteins. 2023. PMID: 36443029 Free PMC article.
-
The 3D-structure, kinetics and dynamics of the E. coli nitroreductase NfsA with NADP+ provide glimpses of its catalytic mechanism.FEBS Lett. 2022 Sep;596(18):2425-2440. doi: 10.1002/1873-3468.14413. Epub 2022 Jul 13. FEBS Lett. 2022. PMID: 35648111 Free PMC article.
-
Flavin oxidation state impacts on nitrofuran antibiotic binding orientation in nitroreductases.Biochem J. 2021 Sep 30;478(18):3423-3428. doi: 10.1042/BCJ20210489. Biochem J. 2021. PMID: 34554213
-
Structure and site-directed mutagenesis of a flavoprotein from Escherichia coli that reduces nitrocompounds: alteration of pyridine nucleotide binding by a single amino acid substitution.J Biol Chem. 2001 Jan 26;276(4):2816-23. doi: 10.1074/jbc.M002617200. Epub 2000 Oct 16. J Biol Chem. 2001. PMID: 11034992
-
Mechanism of CB1954 reduction by Escherichia coli nitroreductase.Biochem Soc Trans. 2009 Apr;37(Pt 2):413-8. doi: 10.1042/BST0370413. Biochem Soc Trans. 2009. PMID: 19290872 Review.
Cited by
-
Oxygen-insensitive nitroreductase E. coli NfsA, but not NfsB, is inhibited by fumarate.Proteins. 2023 May;91(5):585-592. doi: 10.1002/prot.26451. Epub 2022 Dec 13. Proteins. 2023. PMID: 36443029 Free PMC article.
-
Single-cell recordings reveal subpopulations that grow and generate resistance at bactericidal concentrations of antibiotics.Proc Natl Acad Sci U S A. 2023 Feb 14;120(7):e2221546120. doi: 10.1073/pnas.2221546120. Epub 2023 Feb 6. Proc Natl Acad Sci U S A. 2023. PMID: 36745803 Free PMC article. No abstract available.
-
5-Nitrofuran-Tagged Oxazolyl Pyrazolopiperidines: Synthesis and Activity against ESKAPE Pathogens.Molecules. 2023 Sep 7;28(18):6491. doi: 10.3390/molecules28186491. Molecules. 2023. PMID: 37764267 Free PMC article.
-
Evolutionary trajectory of bacterial resistance to antibiotics and antimicrobial peptides in Escherichia coli.mSystems. 2025 Mar 18;10(3):e0170024. doi: 10.1128/msystems.01700-24. Epub 2025 Feb 27. mSystems. 2025. PMID: 40013796 Free PMC article.
-
Genomic insights into plasmid mediated AMR genes, virulence factors and mobile genetic elements in raw milk Escherichia coli from Gujarat, India.Sci Rep. 2025 Feb 21;15(1):6320. doi: 10.1038/s41598-025-90761-2. Sci Rep. 2025. PMID: 39984597 Free PMC article.
References
-
- Whiteway, J., Koziarz, P., Veall, J., Sandhu, N., Kumar, P., Hoecher, B.et al. (1998) Oxygen-insensitive nitroreductases: analysis of the roles of nfsA and nfsB in development of resistance to 5-nitrofuran derivatives in Escherichia coli. J. Bacteriol. 180, 5529–5539 10.1128/JB.180.21.5529-5539.1998 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

