Axial elongation of caudalized human organoids mimics aspects of neural tube development
- PMID: 34142711
- PMCID: PMC8254868
- DOI: 10.1242/dev.198275
Axial elongation of caudalized human organoids mimics aspects of neural tube development
Abstract
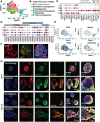
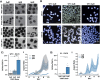
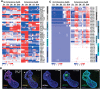
Axial elongation of the neural tube is crucial during mammalian embryogenesis for anterior-posterior body axis establishment and subsequent spinal cord development, but these processes cannot be interrogated directly in humans as they occur post-implantation. Here, we report an organoid model of neural tube extension derived from human pluripotent stem cell (hPSC) aggregates that have been caudalized with Wnt agonism, enabling them to recapitulate aspects of the morphological and temporal gene expression patterns of neural tube development. Elongating organoids consist largely of neuroepithelial compartments and contain TBXT+SOX2+ neuro-mesodermal progenitors in addition to PAX6+NES+ neural progenitors. A critical threshold of Wnt agonism stimulated singular axial extensions while maintaining multiple cell lineages, such that organoids displayed regionalized anterior-to-posterior HOX gene expression with hindbrain (HOXB1) regions spatially distinct from brachial (HOXC6) and thoracic (HOXB9) regions. CRISPR interference-mediated silencing of TBXT, a Wnt pathway target, increased neuroepithelial compartmentalization, abrogated HOX expression and disrupted uniaxial elongation. Together, these results demonstrate the potent capacity of caudalized hPSC organoids to undergo axial elongation in a manner that can be used to dissect the cellular organization and patterning decisions that dictate early human nervous system development.
Keywords: Neural tube development; Organoid models; Stem cell biology.
© 2021. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures





Similar articles
-
Controlling human organoid symmetry breaking reveals signaling gradients drive segmentation clock waves.Cell. 2023 Feb 2;186(3):513-527.e19. doi: 10.1016/j.cell.2022.12.042. Epub 2023 Jan 18. Cell. 2023. PMID: 36657441 Free PMC article.
-
Dynamic 3D Combinatorial Generation of hPSC-Derived Neuromesodermal Organoids With Diverse Regional and Cellular Identities.Curr Protoc. 2022 Oct;2(10):e568. doi: 10.1002/cpz1.568. Curr Protoc. 2022. PMID: 36264199 Free PMC article.
-
Establishing dorsal-ventral patterning in human neural tube organoids with synthetic organizers.Cell Stem Cell. 2025 Jul 3;32(7):1071-1086.e8. doi: 10.1016/j.stem.2025.04.011. Epub 2025 May 14. Cell Stem Cell. 2025. PMID: 40373768
-
Neuromesodermal specification during head-to-tail body axis formation.Curr Top Dev Biol. 2024;159:232-271. doi: 10.1016/bs.ctdb.2024.02.012. Epub 2024 Mar 19. Curr Top Dev Biol. 2024. PMID: 38729677 Review.
-
From Bipotent Neuromesodermal Progenitors to Neural-Mesodermal Interactions during Embryonic Development.Int J Mol Sci. 2021 Aug 24;22(17):9141. doi: 10.3390/ijms22179141. Int J Mol Sci. 2021. PMID: 34502050 Free PMC article. Review.
Cited by
-
Loss of TJP1 disrupts gastrulation patterning and increases differentiation toward the germ cell lineage in human pluripotent stem cells.Dev Cell. 2023 Aug 21;58(16):1477-1488.e5. doi: 10.1016/j.devcel.2023.05.019. Epub 2023 Jun 23. Dev Cell. 2023. PMID: 37354899 Free PMC article.
-
Constructing the pharyngula: Connecting the primary axial tissues of the head with the posterior axial tissues of the tail.Cells Dev. 2023 Dec;176:203866. doi: 10.1016/j.cdev.2023.203866. Epub 2023 Jun 30. Cells Dev. 2023. PMID: 37394035 Free PMC article. Review.
-
Timely TGFβ signalling inhibition induces notochord.Nature. 2025 Jan;637(8046):673-682. doi: 10.1038/s41586-024-08332-w. Epub 2024 Dec 18. Nature. 2025. PMID: 39695233 Free PMC article.
-
Sculpting with stem cells: how models of embryo development take shape.Development. 2021 Dec 15;148(24):dev192914. doi: 10.1242/dev.192914. Epub 2021 Dec 15. Development. 2021. PMID: 34908102 Free PMC article.
-
Neural Tube Organoids: A Novel System to Study Developmental Timing.Stem Cell Rev Rep. 2024 Nov;20(8):2045-2061. doi: 10.1007/s12015-024-10785-5. Epub 2024 Sep 4. Stem Cell Rev Rep. 2024. PMID: 39230820 Free PMC article. Review.
References
-
- Amoroso, M. W., Croft, G. F., Williams, D. J., O'keeffe, S., Carrasco, M. A., Davis, A. R., Roybon, L., Oakley, D. H., Maniatis, T., Henderson, C. E.et al. (2013). Accelerated high-yield generation of limb-innervating motor neurons from human stem cells. J. Neurosci. 33, 574-586. 10.1523/JNEUROSCI.0906-12.2013 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

