Transcriptomic, proteomic and phosphoproteomic underpinnings of daily exercise performance and zeitgeber activity of training in mouse muscle
- PMID: 34142717
- PMCID: PMC9290843
- DOI: 10.1113/JP281535
Transcriptomic, proteomic and phosphoproteomic underpinnings of daily exercise performance and zeitgeber activity of training in mouse muscle
Abstract
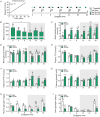
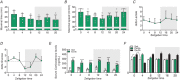
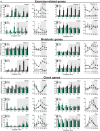
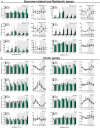
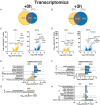
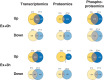
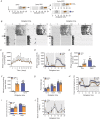
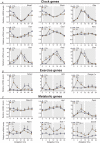
Key points: Maximal endurance performance is greater in the early daytime. Timed exercise differentially alters the muscle transcriptome and (phospho)-proteome. Early daytime exercise triggers energy provisioning and tissue regeneration. Early night-time exercise activates stress-related and catabolic pathways. Scheduled training has limited effects on the muscle and liver circadian clocks.
Abstract: Timed physical activity might potentiate the health benefits of training. The underlying signalling events triggered by exercise at different times of day are, however, poorly understood. Here, we found that time-dependent variations in maximal treadmill exercise capacity of naïve mice were associated with energy stores, mostly hepatic glycogen levels. Importantly, running at different times of day resulted in a vastly different activation of signalling pathways, e.g. related to stress response, vesicular trafficking, repair and regeneration. Second, voluntary wheel running at the opposite phase of the dark, feeding period surprisingly revealed a minimal zeitgeber (i.e. phase-shifting) effect of training on the muscle clock. This integrated study provides important insights into the circadian regulation of endurance performance and the control of the circadian clock by exercise. In future studies, these results could contribute to better understanding circadian aspects of training design in athletes and the application of chrono-exercise-based interventions in patients.
Keywords: circadian clock; energy homeostasis; exercise; metabolism; proteomics; skeletal muscle; transcriptomics; zeitgeber.
© 2021 The Authors. The Journal of Physiology published by John Wiley & Sons Ltd on behalf of The Physiological Society.
Figures





Comment in
-
The muscle clock: how does muscle physiology change in 24 h in response to exercise?J Physiol. 2022 Feb;600(4):701-702. doi: 10.1113/JP282005. Epub 2021 Jul 12. J Physiol. 2022. PMID: 34165195 No abstract available.
-
Circadian control of skeletal muscle adaptations to exercise.J Physiol. 2022 Feb;600(4):723-724. doi: 10.1113/JP282134. Epub 2021 Aug 17. J Physiol. 2022. PMID: 34339519 No abstract available.
-
The many 'coggs' in the food intake clock.J Physiol. 2022 Feb;600(4):703-704. doi: 10.1113/JP282291. Epub 2021 Oct 11. J Physiol. 2022. PMID: 34555176 No abstract available.
References
-
- Ahrne E, Glatter T, Vigano C, Schubert C, Nigg EA & Schmidt A (2016). Evaluation and improvement of quantification accuracy in isobaric mass tag‐based protein quantification experiments. J Proteome Res 15, 2537–2547. - PubMed
-
- Albrecht U & Eichele G (2003). The mammalian circadian clock. Curr Opin Genet Dev 13, 271–277. - PubMed
-
- Allen DL, Harrison BC, Maass A, Bell ML, Byrnes WC & Leinwand LA (2001). Cardiac and skeletal muscle adaptations to voluntary wheel running in the mouse. J Appl Physiol 90, 1900–1908. - PubMed
-
- Bendtsen JD, Jensen LJ, Blom N, Von Heijne G & Brunak S (2004). Feature‐based prediction of non‐classical and leaderless protein secretion. Protein Eng Des Sel 17, 349–356. - PubMed
-
- Boergesen M, Pedersen TA, Gross B, van Heeringen SJ, Hagenbeek D, Bindesboll C, Caron S, Lalloyer F, Steffensen KR, Nebb HI, Gustafsson JA, Stunnenberg HG, Staels B & Mandrup S (2012). Genome‐wide profiling of liver X receptor, retinoid X receptor, and peroxisome proliferator‐activated receptor alpha in mouse liver reveals extensive sharing of binding sites. Mol Cell Biol 32, 852–867. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

