Immunological imprinting of the antibody response in COVID-19 patients
- PMID: 34145263
- PMCID: PMC8213790
- DOI: 10.1038/s41467-021-23977-1
Immunological imprinting of the antibody response in COVID-19 patients
Abstract
In addition to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), humans are also susceptible to six other coronaviruses, for which consecutive exposures to antigenically related and divergent seasonal coronaviruses are frequent. Despite the prevalence of COVID-19 pandemic and ongoing research, the nature of the antibody response against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is unclear. Here we longitudinally profile the early humoral immune response against SARS-CoV-2 in hospitalized coronavirus disease 2019 (COVID-19) patients and quantify levels of pre-existing immunity to OC43, HKU1 and 229E seasonal coronaviruses, and find a strong back-boosting effect to conserved but not variable regions of OC43 and HKU1 betacoronaviruses spike protein. However, such antibody memory boost to human coronaviruses negatively correlates with the induction of IgG and IgM against SARS-CoV-2 spike and nucleocapsid protein. Our findings thus provide evidence of immunological imprinting by previous seasonal coronavirus infections that can potentially modulate the antibody profile to SARS-CoV-2 infection.
Conflict of interest statement
A.G.S. is inventor of patents owned by the Icahn School of Medicine at Mount Sinai in the field of influenza virus vaccines. The A.G.S. lab has received research funds from Avimex, GSK, and 7Hills to investigate novel influenza virus vaccines. The Icahn School of Medicine at Mount Sinai has filed patent applications relating to SARS-CoV-2 serological assays and NDV-based SARS-CoV-2 vaccines which list F.K. as co-inventor. D.S. and F.A. are also listed on the serological assay patent application as co-inventors. Mount Sinai has spun out a company, Kantaro, to market serological tests for SARS-CoV-2. F.K. has consulted for Merck and Pfizer (before 2020), and is currently consulting for Pfizer, Seqirus, and Avimex. The Krammer laboratory is also collaborating with Pfizer on animal models of SARS-CoV-2. The other authors declare no competing interests.
Figures
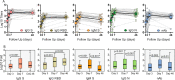
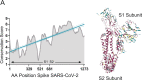




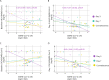

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

