Live-cell imaging of circadian clock protein dynamics in CRISPR-generated knock-in cells
- PMID: 34145278
- PMCID: PMC8213786
- DOI: 10.1038/s41467-021-24086-9
Live-cell imaging of circadian clock protein dynamics in CRISPR-generated knock-in cells
Abstract
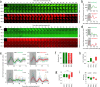
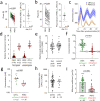
The cell biology of circadian clocks is still in its infancy. Here, we describe an efficient strategy for generating knock-in reporter cell lines using CRISPR technology that is particularly useful for genes expressed transiently or at low levels, such as those coding for circadian clock proteins. We generated single and double knock-in cells with endogenously expressed PER2 and CRY1 fused to fluorescent proteins allowing us to simultaneously monitor the dynamics of CRY1 and PER2 proteins in live single cells. Both proteins are highly rhythmic in the nucleus of human cells with PER2 showing a much higher amplitude than CRY1. Surprisingly, CRY1 protein is nuclear at all circadian times indicating the absence of circadian gating of nuclear import. Furthermore, in the nucleus of individual cells CRY1 abundance rhythms are phase-delayed (~5 hours), and CRY1 levels are much higher (>5 times) compared to PER2 questioning the current model of the circadian oscillator.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

