The FLI portion of EWS/FLI contributes a transcriptional regulatory function that is distinct and separable from its DNA-binding function in Ewing sarcoma
- PMID: 34145397
- PMCID: PMC8298202
- DOI: 10.1038/s41388-021-01876-5
The FLI portion of EWS/FLI contributes a transcriptional regulatory function that is distinct and separable from its DNA-binding function in Ewing sarcoma
Abstract
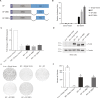
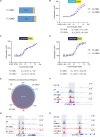
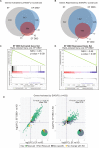
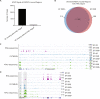
Ewing sarcoma is an aggressive bone cancer of children and young adults defined by the presence of a chromosomal translocation: t(11;22)(q24;q12). The encoded protein, EWS/FLI, fuses the amino-terminal domain of EWS to the carboxyl-terminus of FLI. The EWS portion is an intrinsically disordered transcriptional regulatory domain, while the FLI portion contains an ETS DNA-binding domain and two flanking regions of unknown function. Early studies using non-Ewing sarcoma models provided conflicting information on the roles of each domain of FLI in EWS/FLI oncogenic function. We therefore sought to define the specific contributions of each FLI domain to EWS/FLI activity in a well-validated Ewing sarcoma model and, in doing so, to better understand Ewing sarcoma development mediated by the fusion protein. We analyzed a series of engineered EWS/FLI mutants with alterations in the FLI portion using a variety of assays. Fluorescence anisotropy, CUT&RUN, and ATAC-sequencing experiments revealed that the isolated ETS domain is sufficient to maintain the normal DNA-binding and chromatin accessibility function of EWS/FLI. In contrast, RNA-sequencing and soft agar colony formation assays revealed that the ETS domain alone was insufficient for transcriptional regulatory and oncogenic transformation functions of the fusion protein. We found that an additional alpha-helix immediately downstream of the ETS domain is required for full transcriptional regulation and EWS/FLI-mediated oncogenesis. These data demonstrate a previously unknown role for FLI in transcriptional regulation that is distinct from its DNA-binding activity. This activity is critical for the cancer-causing function of EWS/FLI and may lead to novel therapeutic approaches.
© 2021. The Author(s).
Conflict of interest statement
Research reported in this publication was supported by the National Institute of Health award T32 GM068412 to MAB, and U54 CA231641 to SLL. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. SLL declares a competing interest as a member of the advisory board for Salarius Pharmaceuticals. SLL is also a listed inventor on United States Patent No. US 7,939,253 B2, “Methods and compositions for the diagnosis and treatment of Ewing’s sarcoma,” and United States Patent No. US 8,557,532, “Diagnosis and treatment of drug-resistant Ewing’s sarcoma.” This does not alter our adherence to
Figures


References
-
- Aurias A, Rimbaut C, Buffe D, Dubousset J, Mazabraud A. [Translocation of chromosome 22 in Ewing’s sarcoma] C R Seances Acad Sci III. 1983;296:1105–7. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

