CDK9 keeps RNA polymerase II on track
- PMID: 34146121
- PMCID: PMC8257543
- DOI: 10.1007/s00018-021-03878-8
CDK9 keeps RNA polymerase II on track
Abstract
Cyclin-dependent kinase 9 (CDK9), the kinase component of positive transcription elongation factor b (P-TEFb), is essential for transcription of most protein-coding genes by RNA polymerase II (RNAPII). By releasing promoter-proximally paused RNAPII into gene bodies, CDK9 controls the entry of RNAPII into productive elongation and is, therefore, critical for efficient synthesis of full-length messenger (m)RNAs. In recent years, new players involved in P-TEFb-dependent processes have been identified and an important function of CDK9 in coordinating elongation with transcription initiation and termination has been unveiled. As the regulatory functions of CDK9 in gene expression continue to expand, a number of human pathologies, including cancers, have been associated with aberrant CDK9 activity, underscoring the need to properly regulate CDK9. Here, I provide an overview of CDK9 function and regulation, with an emphasis on CDK9 dysregulation in human diseases.
Keywords: 7SK RNA; Cyclin T1; HIV; Promoter-proximal pausing; RNA polymerase II CTD; Transcriptional checkpoint.
Conflict of interest statement
The author declares no competing interests.
Figures

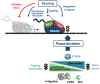

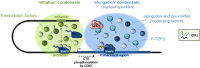

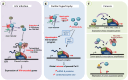
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

