KDELR2 promotes breast cancer proliferation via HDAC3-mediated cell cycle progression
- PMID: 34146461
- PMCID: PMC8441056
- DOI: 10.1002/cac2.12180
KDELR2 promotes breast cancer proliferation via HDAC3-mediated cell cycle progression
Abstract
Background: Histone deacetylases (HDACs) engage in the regulation of various cellular processes by controlling global gene expression. The dysregulation of HDACs leads to carcinogenesis, making HDACs ideal targets for cancer therapy. However, the use of HDAC inhibitors (HDACi) as single agents has been shown to have limited success in treating solid tumors in clinical studies. This study aimed to identify a novel downstream effector of HDACs to provide a potential target for combination therapy.
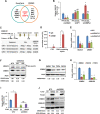
Methods: Transcriptome sequencing and bioinformatics analysis were performed to screen for genes responsive to HDACi in breast cancer cells. The effects of HDACi on cell viability were detected using the MTT assay. The mRNA and protein levels of genes were determined by quantitative reverse transcription-PCR (qRT-PCR) and Western blotting. Cell cycle distribution and apoptosis were analyzed by flow cytometry. The binding of CREB1 (cAMP-response element binding protein 1) to the promoter of the KDELR (The KDEL (Lys-Asp-Glu-Leu) receptor) gene was validated by the ChIP (chromatin immunoprecipitation assay). The association between KDELR2 and protein of centriole 5 (POC5) was detected by immunoprecipitation. A breast cancer-bearing mouse model was employed to analyze the effect of the HDAC3-KDELR2 axis on tumor growth.
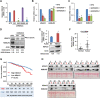
Results: KDELR2 was identified as a novel target of HDAC3, and its aberrant expression indicated the poor prognosis of breast cancer patients. We found a strong correlation between the protein expression patterns of HADC3 and KDELR2 in tumor tissues from breast cancer patients. The results of the ChIP assay and qRT-PCR analysis validated that HDAC3 transactivated KDELR2 via CREB1. The HDAC3-KDELR2 axis accelerated the cell cycle progression of cancer cells by protecting the centrosomal protein POC5 from proteasomal degradation. Moreover, the HDAC3-KDELR2 axis promoted breast cancer cell proliferation and tumorigenesis in vitro and in vivo.
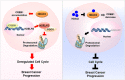
Conclusion: Our results uncovered a previously unappreciated function of KDELR2 in tumorigenesis, linking a critical Golgi-the endoplasmic reticulum traffic transport protein to HDAC-controlled cell cycle progression on the path of cancer development and thus revealing a potential therapeutical target for breast cancer.
Keywords: CREB1; HADC3; KDELR2; breast cancer; histone deacetylase inhibitor; protein of centriole 5.
© 2021 The Authors. Cancer Communications published by John Wiley & Sons Australia, Ltd. on behalf of Sun Yat-sen University Cancer Center.
Figures



References
-
- Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150(1):12–27. - PubMed
-
- Marks P, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. Histone deacetylases and cancer: causes and therapies. Nat Rev Cancer. 2001;1(3):194–202. - PubMed
-
- Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov. 2014;13(9):673–91. - PubMed
-
- Romero D. HDAC inhibitors tested in phase III trial. Nat Rev Clin Oncol. 2019;16(8):465‐. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

