Mucosal Biofilms Are an Endoscopic Feature of Irritable Bowel Syndrome and Ulcerative Colitis
- PMID: 34146566
- PMCID: PMC8527885
- DOI: 10.1053/j.gastro.2021.06.024
Mucosal Biofilms Are an Endoscopic Feature of Irritable Bowel Syndrome and Ulcerative Colitis
Abstract
Background & aims: Irritable bowel syndrome (IBS) and inflammatory bowel diseases result in a substantial reduction in quality of life and a considerable socioeconomic impact. In IBS, diagnosis and treatment options are limited, but evidence for involvement of the gut microbiome in disease pathophysiology is emerging. Here we analyzed the prevalence of endoscopically visible mucosal biofilms in gastrointestinal disease and associated changes in microbiome composition and metabolism.
Methods: The presence of mucosal biofilms was assessed in 1426 patients at 2 European university-based endoscopy centers. One-hundred and seventeen patients were selected for in-depth molecular and microscopic analysis using 16S ribosomal RNA gene amplicon-sequencing of colonic biopsies and fecal samples, confocal microscopy with deep learning-based image analysis, scanning electron microscopy, metabolomics, and in vitro biofilm formation assays.
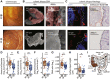
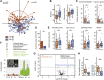
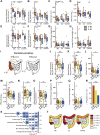
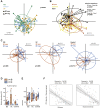
Results: Biofilms were present in 57% of patients with IBS and 34% of patients with ulcerative colitis compared with 6% of controls (P < .001). These yellow-green adherent layers of the ileum and right-sided colon were microscopically confirmed to be dense bacterial biofilms. 16S-sequencing links the presence of biofilms to a dysbiotic gut microbiome, including overgrowth of Escherichia coli and Ruminococcus gnavus. R. gnavus isolates cultivated from patient biofilms also formed biofilms in vitro. Metabolomic analysis found an accumulation of bile acids within biofilms that correlated with fecal bile acid excretion, linking this phenotype with a mechanism of diarrhea.
Conclusions: The presence of mucosal biofilms is an endoscopic feature in a subgroup of IBS and ulcerative colitis with disrupted bile acid metabolism and bacterial dysbiosis. They provide novel insight into the pathophysiology of IBS and ulcerative colitis, illustrating that biofilm can be seen as a tipping point in the development of dysbiosis and disease.
Keywords: Bacterial–Epithelial Interaction; Endoscopy; Functional Gastrointestinal Disorders; Microbiota.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Figures









Comment in
-
PEG and Mucosal Biofilms in Irritable Bowel Syndrome and Ulcerative Colitis.Gastroenterology. 2022 Mar;162(3):992-993. doi: 10.1053/j.gastro.2021.06.053. Epub 2021 Jun 25. Gastroenterology. 2022. PMID: 34175284 No abstract available.
-
MUW researcher of the month.Wien Klin Wochenschr. 2022 Mar;134(5-6):255-257. doi: 10.1007/s00508-022-02023-0. Wien Klin Wochenschr. 2022. PMID: 35286483 No abstract available.
References
Supplementary References
-
- Swidsinski A., Loening-Baucke V., Bengmark S. Bacterial Biofilm suppression with antibiotics for ulcerative and indeterminate colitis: consequences of aggressive treatment. Archives of Medical Research. 2008;39:198–204. - PubMed
-
- Brown M.R., Gilbert P. Sensitivity of biofilms to antimicrobial agents. J Appl Bacteriol. 1993;74(Suppl):87S–97S. - PubMed
-
- Falk T., Mai D., Bensch R. U-Net: deep learning for cell counting, detection, and morphometry. Nat Methods. 2019;16:67–70. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

