Enhanced HSC-like cell generation from mouse pluripotent stem cells in a 3D induction system cocultured with stromal cells
- PMID: 34147128
- PMCID: PMC8214308
- DOI: 10.1186/s13287-021-02434-2
Enhanced HSC-like cell generation from mouse pluripotent stem cells in a 3D induction system cocultured with stromal cells
Abstract
Background: Decades of efforts have attempted to differentiate the pluripotent stem cells (PSCs) into truly functional hematopoietic stem cells (HSCs), yet the problems of low differentiation efficiency in vitro and poor hematopoiesis reconstitution in vivo still exist, mainly attributing to the lack of solid, reproduced, or pursued differentiation system.
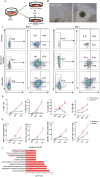
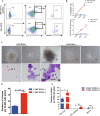
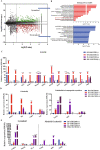
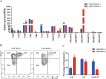
Methods: In this study, we established an in vitro differentiation system yielding in vivo hematopoietic reconstitution hematopoietic cells from mouse PSCs through a 3D induction system followed by coculture with OP9 stromal cells. The in vivo hematopoietic reconstitution potential of c-kit+ cells derived from the mouse PSCs was evaluated via m-NSG transplantation assay. Flow cytometry analysis, RNA-seq, and cell cycle analysis were used to detect the in vitro hematopoietic ability of endothelial protein C receptor (EPCR, CD201) cells generated in our induction system.
Results: The c-kit+ cells from 3D self-assembling peptide induction system followed by the OP9 coculture system possessed apparently superiority in terms of in vivo repopulating activity than that of 3D induction system followed by the 0.1% gelatin culture. We interestingly found that our 3D+OP9 system enriched a higher percentage of CD201+c-kit+cells that showed more similar HSC-like features such as transcriptome level and CFU formation ability than CD201-c-kit+cells, which have not been reported in the field of mouse PSCs hematopoietic differentiation. Moreover, CD201+ hematopoietic cells remained in a relatively slow cycling state, consistent with high expression levels of P57 and Ccng2. Further, we innovatively demonstrated that notch signaling pathway is responsible for in vitro CD201+ hematopoietic cell induction from mouse PSCs.
Conclusions: Altogether, our findings lay a foundation for improving the efficiency of hematopoietic differentiation and generating in vivo functional HSC-like cells from mouse PSCs for clinical application.
Keywords: 3D system; CD201; Hematopoiesis; Notch; Pluripotent stem cells.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Wahlster L, Daley GJN. Progress towards generation of human haematopoietic stem cells. Nat Cell Biol. 2016;18(11):1111–7. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

