Identification of sixteen novel candidate genes for late onset Parkinson's disease
- PMID: 34148545
- PMCID: PMC8215754
- DOI: 10.1186/s13024-021-00455-2
Identification of sixteen novel candidate genes for late onset Parkinson's disease
Abstract
Background: Parkinson's disease (PD) is a neurodegenerative movement disorder affecting 1-5% of the general population for which neither effective cure nor early diagnostic tools are available that could tackle the pathology in the early phase. Here we report a multi-stage procedure to identify candidate genes likely involved in the etiopathogenesis of PD.
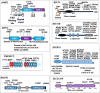
Methods: The study includes a discovery stage based on the analysis of whole exome data from 26 dominant late onset PD families, a validation analysis performed on 1542 independent PD patients and 706 controls from different cohorts and the assessment of polygenic variants load in the Italian cohort (394 unrelated patients and 203 controls).
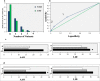
Results: Family-based approach identified 28 disrupting variants in 26 candidate genes for PD including PARK2, PINK1, DJ-1(PARK7), LRRK2, HTRA2, FBXO7, EIF4G1, DNAJC6, DNAJC13, SNCAIP, AIMP2, CHMP1A, GIPC1, HMOX2, HSPA8, IMMT, KIF21B, KIF24, MAN2C1, RHOT2, SLC25A39, SPTBN1, TMEM175, TOMM22, TVP23A and ZSCAN21. Sixteen of them have not been associated to PD before, were expressed in mesencephalon and were involved in pathways potentially deregulated in PD. Mutation analysis in independent cohorts disclosed a significant excess of highly deleterious variants in cases (p = 0.0001), supporting their role in PD. Moreover, we demonstrated that the co-inheritance of multiple rare variants (≥ 2) in the 26 genes may predict PD occurrence in about 20% of patients, both familial and sporadic cases, with high specificity (> 93%; p = 4.4 × 10- 5). Moreover, our data highlight the fact that the genetic landmarks of late onset PD does not systematically differ between sporadic and familial forms, especially in the case of small nuclear families and underline the importance of rare variants in the genetics of sporadic PD. Furthermore, patients carrying multiple rare variants showed higher risk of manifesting dyskinesia induced by levodopa treatment.
Conclusions: Besides confirming the extreme genetic heterogeneity of PD, these data provide novel insights into the genetic of the disease and may be relevant for its prediction, diagnosis and treatment.
Keywords: Late onset Parkinson’s disease; Novel candidate genes for Parkinson’s disease; Rare variant burden analysis; Whole exome sequencing.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Jellinger KA. The pathology of Parkinson’s disease. Adv Neurol. 2001;86:55–72. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

