Programmed Deviations of Ribosomes From Standard Decoding in Archaea
- PMID: 34149676
- PMCID: PMC8211752
- DOI: 10.3389/fmicb.2021.688061
Programmed Deviations of Ribosomes From Standard Decoding in Archaea
Abstract
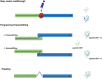
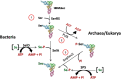
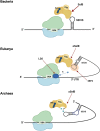
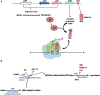
Genetic code decoding, initially considered to be universal and immutable, is now known to be flexible. In fact, in specific genes, ribosomes deviate from the standard translational rules in a programmed way, a phenomenon globally termed recoding. Translational recoding, which has been found in all domains of life, includes a group of events occurring during gene translation, namely stop codon readthrough, programmed ± 1 frameshifting, and ribosome bypassing. These events regulate protein expression at translational level and their mechanisms are well known and characterized in viruses, bacteria and eukaryotes. In this review we summarize the current state-of-the-art of recoding in the third domain of life. In Archaea, it was demonstrated and extensively studied that translational recoding regulates the decoding of the 21st and the 22nd amino acids selenocysteine and pyrrolysine, respectively, and only one case of programmed -1 frameshifting has been reported so far in Saccharolobus solfataricus P2. However, further putative events of translational recoding have been hypothesized in other archaeal species, but not extensively studied and confirmed yet. Although this phenomenon could have some implication for the physiology and adaptation of life in extreme environments, this field is still underexplored and genes whose expression could be regulated by recoding are still poorly characterized. The study of these recoding episodes in Archaea is urgently needed.
Keywords: alpha-fucosidase; archaea; frameshifting; pyrrolysine; recoding; selenocysteine.
Copyright © 2021 De Lise, Strazzulli, Iacono, Curci, Di Fenza, Maurelli, Moracci and Cobucci-Ponzano.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources

