Agrobacterium VirE2 Protein Modulates Plant Gene Expression and Mediates Transformation From Its Location Outside the Nucleus
- PMID: 34149784
- PMCID: PMC8213393
- DOI: 10.3389/fpls.2021.684192
Agrobacterium VirE2 Protein Modulates Plant Gene Expression and Mediates Transformation From Its Location Outside the Nucleus
Abstract
Agrobacterium effector protein VirE2 is important for plant transformation. VirE2 likely coats transferred DNA (T-DNA) in the plant cell and protects it from degradation. VirE2 localizes to the plant cytoplasm and interacts with several host proteins. Plant-expressed VirE2 can complement a virE2 mutant Agrobacterium strain to support transformation. We investigated whether VirE2 could facilitate transformation from a nuclear location by affixing to it a strong nuclear localization signal (NLS) sequence. Only cytoplasmic-, but not nuclear-localized, VirE2 could stimulate transformation. To investigate the ways VirE2 supports transformation, we generated transgenic Arabidopsis plants containing a virE2 gene under the control of an inducible promoter and performed RNA-seq and proteomic analyses before and after induction. Some differentially expressed plant genes were previously known to facilitate transformation. Knockout mutant lines of some other VirE2 differentially expressed genes showed altered transformation phenotypes. Levels of some proteins known to be important for transformation increased in response to VirE2 induction, but prior to or without induction of their corresponding mRNAs. Overexpression of some other genes whose proteins increased after VirE2 induction resulted in increased transformation susceptibility. We conclude that cytoplasmically localized VirE2 modulates both plant RNA and protein levels to facilitate transformation.
Keywords: Agrobacterium; Arabidopsis; VirE2; plant transformation; protein subcellular localization; proteome; transcriptome.
Copyright © 2021 Lapham, Lee, Xhako, Gómez, Nivya and Gelvin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
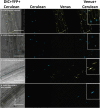
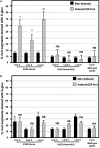


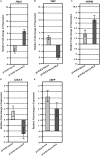

References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

