Tailoring strain construction strategies for muconic acid production in S. cerevisiae and E. coli
- PMID: 34150501
- PMCID: PMC8193250
- DOI: 10.1016/j.meteno.2014.09.001
Tailoring strain construction strategies for muconic acid production in S. cerevisiae and E. coli
Abstract
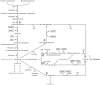
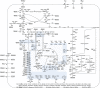
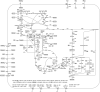
There is currently a strong interest to derive the biological precursor cis,cis-muconic acid from shikimate pathway-branches to develop a biological replacement for adipic acid. Pioneered by the Frost laboratory this concept has regained interest: Recent approaches (Boles, Alper, Yan) however suffer from low product titres. Here an in silico comparison of all strain construction strategies was conducted to highlight stoichiometric optimizations. Using elementary mode analysis new knock-out strategies were determined in Saccharomyces cerevisiae and Escherichia coli. The strain construction strategies are unique to each pathway-branch and organism, allowing significantly different maximum and minimum yields. The maximum theoretical product carbon yields on glucose ranged from 86% (dehydroshikimate-branch) to 69% (anthranilate-branch). In most cases a coupling of product formation to growth was possible. Especially in S. cerevisiae chorismate-routes a minimum yield constraint of 46.9% could be reached. The knock-out targets are non-obvious, and not-transferable, highlighting the importance of tailored strain construction strategies.
Keywords: ADH, acetaldehyde dehydrogenase; ANTH, anthranilate; Adipic; Carbon yield; DAHP, 3-deoxy-arabinoheptulosonate 7-phosphate; DHBA, 2,3-dihydroxybenzoate; DHS, 3-dehydroshikimate; E4P, erythrose 4-phosphate; EDA, 2-dehydro-3-deoxy-phosphogluconate aldolase; EDP, Entner–Doudoroff pathway; EFM, elementary flux modes; EMA, elementary mode analysis; EPSP, 5-enolpyruvylshikimate-3-phosphate; Elementary mode analysis; F6P, fructose-6-phosphate; FRD, fumarate reductase; G6PD, glucose-6-phosphate dehydrogenase; GPDH, glycerol-3-phosphate dehydrogenase; Knock-out strategies; LDH, lactate dehydrogenase; MAE, malic enzyme; Muconic; PCA, protocatechuate; PCK, phosphoenolpyruvate carboxykinase; PEP, phosphoenolpyruvate; PPP, pentose phosphate pathway; PPS, phosphoenolpyruvate synthase; PTS, phosphotransferase system; PYK, pyruvate kinase; SA, salicylate; WT, wild type; Ymax, maximum theoretical carbon yield; Ymin, minimum theoretical carbon yield; cMCs, constrained minimal cut sets; ccMA, cis,cis-muconic acid; pHBA, para-hydroxybenzoate.
© 2014 The Authors.
Figures


References
-
- Alini S., Basile F., Blasioli S., Rinaldi C., Vaccari A. Development of new catalysts for N2O-decomposition from adipic acid plant. Appl. Catal. B Environ. 2007;70:323–329.
-
- Baneyx F. Recombinant protein expression in Escherichia coli. Curr. Opin. Biotechnol. 1999;10:411–421. - PubMed
-
- Bang S.-G., Choi C.Y. DO-stat fed-batch production of cis,cis-muconic acid from benzoic acid by Pseudomonas putida BM014. J. Ferment. Bioeng. 1995;79:381–383.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

