CRISPR-Cas system enables fast and simple genome editing of industrial Saccharomyces cerevisiae strains
- PMID: 34150504
- PMCID: PMC8193243
- DOI: 10.1016/j.meteno.2015.03.001
CRISPR-Cas system enables fast and simple genome editing of industrial Saccharomyces cerevisiae strains
Abstract
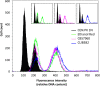
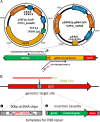
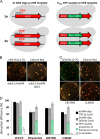
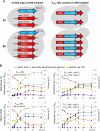
There is a demand to develop 3rd generation biorefineries that integrate energy production with the production of higher value chemicals from renewable feedstocks. Here, robust and stress-tolerant industrial strains of Saccharomyces cerevisiae will be suitable production organisms. However, their genetic manipulation is challenging, as they are usually diploid or polyploid. Therefore, there is a need to develop more efficient genetic engineering tools. We applied a CRISPR-Cas9 system for genome editing of different industrial strains, and show simultaneous disruption of two alleles of a gene in several unrelated strains with the efficiency ranging between 65% and 78%. We also achieved simultaneous disruption and knock-in of a reporter gene, and demonstrate the applicability of the method by designing lactic acid-producing strains in a single transformation event, where insertion of a heterologous gene and disruption of two endogenous genes occurred simultaneously. Our study provides a foundation for efficient engineering of industrial yeast cell factories.
Keywords: Biorefineries; CRISPR–Cas9; CRISPR–Cas9, clustered regularly interspaced short palindromic repeats–CRISPR-associated endonuclease 9; Chemical production; DSB, double strand break; GOI, gene of interest; Genome editing; HDR, homology-directed repair; HR, homologous recombination; Industrial yeast; NHEJ, non-homologous end joining; PAM, protospacer adjacent motif; PI, propidium iodide; SNPs, single nucleotide polymorphisms; TALENs, transcription activator-like effector nucleases; USER, uracil-specific excision reaction; ZFNs, zinc finger nucleases; crRNA, CRISPR RNA; gRNA, guide RNA; tracrRNA, trans-activating RNA.
© 2015 The Authors.
Figures
References
-
- Achilli A., Matmati N., Casalone E., Morpurgo G., Lucaccioni A., Pavlov Y.I., Babudri N. The exceptionally high rate of spontaneous mutations in the polymerase delta proofreading exonuclease-deficient Saccharomyces cerevisiae strain starved for adenine. BMC Genet. 2004;5:7429–7437. doi: 10.1186/1471-2156-5-34. - DOI - PMC - PubMed
-
- Borodina I., Kildegaard K.R., Jensen N.B., Blicher T.H., Maury J., Sherstyk S., Schneider K., Lamosa P., Herrgård M.J., Rosenstand I., Öberg F., Forster J., Nielsen J. Establishing a synthetic pathway for high-level production of 3-hydroxypropionic acid in Saccharomyces cerevisiae via β-alanine. Metab. Eng. 2015;27:57–64. doi: 10.1016/j.ymben.2014.10.003. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

