Protein and Microbial Biomarkers in Sputum Discern Acute and Latent Tuberculosis in Investigation of Pastoral Ethiopian Cohort
- PMID: 34150670
- PMCID: PMC8212885
- DOI: 10.3389/fcimb.2021.595554
Protein and Microbial Biomarkers in Sputum Discern Acute and Latent Tuberculosis in Investigation of Pastoral Ethiopian Cohort
Abstract
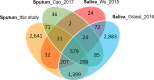
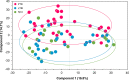
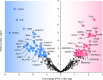
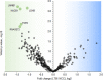
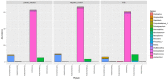
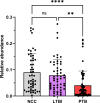
Differential diagnosis of tuberculosis (TB) and latent TB infection (LTBI) remains a public health priority in high TB burden countries. Pulmonary TB is diagnosed by sputum smear microscopy, chest X-rays, and PCR tests for distinct Mycobacterium tuberculosis (Mtb) genes. Clinical tests to diagnose LTBI rely on immune cell stimulation in blood plasma with TB-specific antigens followed by measurements of interferon-γ concentrations. The latter is an important cytokine for cellular immune responses against Mtb in infected lung tissues. Sputum smear microscopy and chest X-rays are not sufficiently sensitive while both PCR and interferon-γ release assays are expensive. Alternative biomarkers for the development of diagnostic tests to discern TB disease states are desirable. This study's objective was to discover sputum diagnostic biomarker candidates from the analysis of samples from 161 human subjects including TB patients, individuals with LTBI, negative community controls (NCC) from the province South Omo, a pastoral region in Ethiopia. We analyzed 16S rRNA gene-based bacterial taxonomies and proteomic profiles. The sputum microbiota did not reveal statistically significant differences in α-diversity comparing the cohorts. The genus Mycobacterium, representing Mtb, was only identified for the TB group which also featured reduced abundance of the genus Rothia in comparison with the LTBI and NCC groups. Rothia is a respiratory tract commensal and may be sensitive to the inflammatory milieu generated by infection with Mtb. Proteomic data supported innate immune responses against the pathogen in subjects with pulmonary TB. Ferritin, an iron storage protein released by damaged host cells, was markedly increased in abundance in TB sputum compared to the LTBI and NCC groups, along with the α-1-acid glycoproteins ORM1 and ORM2. These proteins are acute phase reactants and inhibit excessive neutrophil activation. Proteomic data highlight the effector roles of neutrophils in the anti-Mtb response which was not observed for LTBI cases. Less abundant in the sputum of the LTBI group, compared to the NCC group, were two immunomodulatory proteins, mitochondrial TSPO and the extracellular ribonuclease T2. If validated, these proteins are of interest as new biomarkers for diagnosis of LTBI.
Keywords: LTBI; Rothia; acute phase response; microbiome; protein biomarker; sputum; tuberculosis; α-1-acid glycoprotein.
Copyright © 2021 HaileMariam, Yu, Singh, Teklu, Wondale, Worku, Zewude, Mounaud, Tsitrin, Legesse, Gobena and Pieper.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Differences in plasma proteomes for active tuberculosis, latent tuberculosis and non-tuberculosis mycobacterial lung disease patients with and without ESAT-6/CFP10 stimulation.Proteome Sci. 2020 Oct 31;18(1):10. doi: 10.1186/s12953-020-00165-5. Proteome Sci. 2020. PMID: 33292280 Free PMC article.
-
Algorithms for Screening for Active Tuberculosis among Individuals with Latent Tuberculosis Infection in a Rural Community in China.Microbiol Spectr. 2022 Dec 21;10(6):e0296722. doi: 10.1128/spectrum.02967-22. Epub 2022 Nov 29. Microbiol Spectr. 2022. PMID: 36445141 Free PMC article.
-
Cell-Mediated Immune Responses to in vivo-Expressed and Stage-Specific Mycobacterium tuberculosis Antigens in Latent and Active Tuberculosis Across Different Age Groups.Front Immunol. 2020 Feb 11;11:103. doi: 10.3389/fimmu.2020.00103. eCollection 2020. Front Immunol. 2020. PMID: 32117257 Free PMC article.
-
[Characteristics of a diagnostic method for tuberculosis infection based on whole blood interferon-gamma assay].Kekkaku. 2006 Nov;81(11):681-6. Kekkaku. 2006. PMID: 17154047 Review. Japanese.
-
From immunology to artificial intelligence: revolutionizing latent tuberculosis infection diagnosis with machine learning.Mil Med Res. 2023 Nov 28;10(1):58. doi: 10.1186/s40779-023-00490-8. Mil Med Res. 2023. PMID: 38017571 Free PMC article. Review.
Cited by
-
Plasma Ribonuclease Activity in Antiretroviral Treatment-Naive People With Human Immunodeficiency Virus and Tuberculosis Disease.J Infect Dis. 2024 Aug 16;230(2):403-410. doi: 10.1093/infdis/jiae143. J Infect Dis. 2024. PMID: 38526179 Free PMC article.
-
HBFP: a new repository for human body fluid proteome.Database (Oxford). 2021 Oct 13;2021:baab065. doi: 10.1093/database/baab065. Database (Oxford). 2021. PMID: 34642750 Free PMC article.
-
Predictive biomarkers for latent Mycobacterium tuberculosis infection.Tuberculosis (Edinb). 2024 Jul;147:102399. doi: 10.1016/j.tube.2023.102399. Epub 2023 Aug 24. Tuberculosis (Edinb). 2024. PMID: 37648595 Free PMC article.
-
Dynamic changes of respiratory microbiota associated with treatment outcome in drug-sensitive and drug-resistant pulmonary tuberculosis.Ann Clin Microbiol Antimicrob. 2024 Sep 9;23(1):83. doi: 10.1186/s12941-024-00742-y. Ann Clin Microbiol Antimicrob. 2024. PMID: 39252020 Free PMC article.
-
Genome-wide expression in human whole blood for diagnosis of latent tuberculosis infection: a multicohort research.Front Microbiol. 2025 May 9;16:1584360. doi: 10.3389/fmicb.2025.1584360. eCollection 2025. Front Microbiol. 2025. PMID: 40415930 Free PMC article.
References
-
- Bastian I., Coulter C., National Tuberculosis Advisory C (2017). Position statement on interferon-gamma release assays for the detection of latent tuberculosis infection. Commun. Dis. Intell. Q Rep. 41, E322–EE36. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

