Network-based systems pharmacology reveals heterogeneity in LCK and BCL2 signaling and therapeutic sensitivity of T-cell acute lymphoblastic leukemia
- PMID: 34151288
- PMCID: PMC8208590
- DOI: 10.1038/s43018-020-00167-4
Network-based systems pharmacology reveals heterogeneity in LCK and BCL2 signaling and therapeutic sensitivity of T-cell acute lymphoblastic leukemia
Abstract
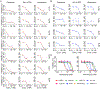
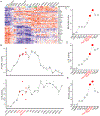
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematological malignancy, and novel therapeutics are much needed. Profiling patient leukemia' drug sensitivities ex vivo, we discovered that 44.4% of childhood and 16.7% of adult T-ALL cases exquisitely respond to dasatinib. Applying network-based systems pharmacology analyses to examine signal circuitry, we identified preTCR-LCK activation as the driver of dasatinib sensitivity, and T-ALL-specific LCK dependency was confirmed in genome-wide CRISPR-Cas9 screens. Dasatinib-sensitive T-ALLs exhibited high BCL-XL and low BCL2 activity and venetoclax resistance. Discordant sensitivity of T-ALL to dasatinib and venetoclax is strongly correlated with T-cell differentiation, particularly with the dynamic shift in LCK vs. BCL2 activation. Finally, single-cell analysis identified leukemia heterogeneity in LCK and BCL2 signaling and T-cell maturation stage, consistent with dasatinib response. In conclusion, our results indicate that developmental arrest in T-ALL drives differential activation of preTCR-LCK and BCL2 signaling in this leukemia, providing unique opportunities for targeted therapy.
Figures










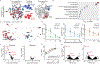




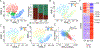
Comment in
-
Targeting the developmental origins of cancer.Nat Cancer. 2021 Mar;2(3):256-257. doi: 10.1038/s43018-021-00186-9. Nat Cancer. 2021. PMID: 35121958 No abstract available.
References
-
- Hunger SP & Mullighan CG Acute lymphoblastic leukemia in children. N Engl J Med 373, 1541–52 (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

