Serological analysis reveals an imbalanced IgG subclass composition associated with COVID-19 disease severity
- PMID: 34151306
- PMCID: PMC8205277
- DOI: 10.1016/j.xcrm.2021.100329
Serological analysis reveals an imbalanced IgG subclass composition associated with COVID-19 disease severity
Abstract
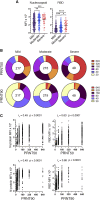
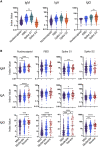
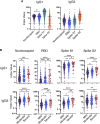
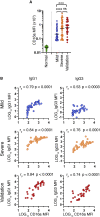
Coronavirus disease 2019 (COVID-19) is associated with a wide spectrum of disease presentation, ranging from asymptomatic infection to acute respiratory distress syndrome (ARDS). Paradoxically, a direct relationship has been suggested between COVID-19 disease severity and the levels of circulating severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-specific antibodies, including virus-neutralizing titers. A serological analysis of 536 convalescent healthcare workers reveals that SARS-CoV-2-specific and virus-neutralizing antibody levels are elevated in individuals that experience severe disease. The severity-associated increase in SARS-CoV-2-specific antibody is dominated by immunoglobulin G (IgG), with an IgG subclass ratio skewed toward elevated receptor binding domain (RBD)- and S1-specific IgG3. In addition, individuals that experience severe disease show elevated SARS-CoV-2-specific antibody binding to the inflammatory receptor FcɣRIIIa. Based on these correlational studies, we propose that spike-specific IgG subclass utilization may contribute to COVID-19 disease severity through potent Fc-mediated effector functions. These results may have significant implications for SARS-CoV-2 vaccine design and convalescent plasma therapy.
Keywords: COVID-19; Fc-effector functions; IgG subclass; SARS-CoV-2; serology.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- McLaughlin C.C., Doll M.K., Morrison K.T., McLaughlin W.L., O’Connor T., Sholukh A.M., Bossard E.L., Phasouk K., Ford E.S., Diem K. High community SARS-CoV-2 antibody seroprevalence in a ski resort community, Blaine County, Idaho, US. preliminary results. medRxiv. 2020 doi: 10.1101/2020.07.19.20157198. - DOI
-
- Pollán M., Pérez-Gómez B., Pastor-Barriuso R., Oteo J., Hernán M.A., Pérez-Olmeda M., Sanmartín J.L., Fernández-García A., Cruz I., Fernández de Larrea N., ENE-COVID Study Group Prevalence of SARS-CoV-2 in Spain (ENE-COVID): a nationwide, population-based seroepidemiological study. Lancet. 2020;396:535–544. - PMC - PubMed
-
- Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D’Andrea K., UPenn COVID Processing Unit Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:eabc8511. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

