Long Reads Are Revolutionizing 20 Years of Insect Genome Sequencing
- PMID: 34152413
- PMCID: PMC8358217
- DOI: 10.1093/gbe/evab138
Long Reads Are Revolutionizing 20 Years of Insect Genome Sequencing
Abstract
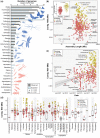
The first insect genome assembly (Drosophila melanogaster) was published two decades ago. Today, nuclear genome assemblies are available for a staggering 601 insect species representing 20 orders. In this study, we analyzed the most-contiguous assembly for each species and provide a "state-of-the-field" perspective, emphasizing taxonomic representation, assembly quality, gene completeness, and sequencing technologies. Relative to species richness, genomic efforts have been biased toward four orders (Diptera, Hymenoptera, Collembola, and Phasmatodea), Coleoptera are underrepresented, and 11 orders still lack a publicly available genome assembly. The average insect genome assembly is 439.2 Mb in length with 87.5% of single-copy benchmarking genes intact. Most notable has been the impact of long-read sequencing; assemblies that incorporate long reads are ∼48× more contiguous than those that do not. We offer four recommendations as we collectively continue building insect genome resources: 1) seek better integration between independent research groups and consortia, 2) balance future sampling between filling taxonomic gaps and generating data for targeted questions, 3) take advantage of long-read sequencing technologies, and 4) expand and improve gene annotations.
Keywords: Arthropoda; Insecta; Oxford Nanopore; Pacific Biosciences; arthropod genomics; long-read sequencing.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

References
-
- Adams MD, et al.2000. The genome sequence of Drosophila melanogaster. Science 287:2185–2195. - PubMed
-
- Bellinger PF, Christiansen KA, Janssens F.. 2020. Checklist of the Collembola of the world. Available from: http://www.collembola.org.
-
- Collins FS, Morgan M, Patrinos A.. 2003. The Human Genome Project: lessons from large-scale biology. Science 300(5617):286–290. - PubMed

