Cluster of Oseltamivir-Resistant and Hemagglutinin Antigenically Drifted Influenza A(H1N1)pdm09 Viruses, Texas, USA, January 2020
- PMID: 34152954
- PMCID: PMC8237887
- DOI: 10.3201/eid2707.204593
Cluster of Oseltamivir-Resistant and Hemagglutinin Antigenically Drifted Influenza A(H1N1)pdm09 Viruses, Texas, USA, January 2020
Abstract
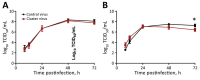
Four cases of oseltamivir-resistant influenza A(H1N1)pdm09 virus infection were detected among inhabitants of a border detention center in Texas, USA. Hemagglutinin of these viruses belongs to 6B.1A5A-156K subclade, which may enable viral escape from preexisting immunity. Our finding highlights the necessity to monitor both drug resistance and antigenic drift of circulating viruses.
Keywords: Texas; United States; antigenic drift; antimicrobial resistance; drug resistance; influenza; influenza A(H1N1); neuraminidase inhibitor; transmission; viruses.
Figures

References
-
- Wu WL, Lau SY, Chen Y, Wang G, Mok BW, Wen X, et al. The 2008-2009 H1N1 influenza virus exhibits reduced susceptibility to antibody inhibition: Implications for the prevalence of oseltamivir resistant variant viruses. Antiviral Res. 2012;93:144–53. 10.1016/j.antiviral.2011.11.006 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

