pyTFM: A tool for traction force and monolayer stress microscopy
- PMID: 34153027
- PMCID: PMC8248623
- DOI: 10.1371/journal.pcbi.1008364
pyTFM: A tool for traction force and monolayer stress microscopy
Abstract
Cellular force generation and force transmission are of fundamental importance for numerous biological processes and can be studied with the methods of Traction Force Microscopy (TFM) and Monolayer Stress Microscopy. Traction Force Microscopy and Monolayer Stress Microscopy solve the inverse problem of reconstructing cell-matrix tractions and inter- and intra-cellular stresses from the measured cell force-induced deformations of an adhesive substrate with known elasticity. Although several laboratories have developed software for Traction Force Microscopy and Monolayer Stress Microscopy computations, there is currently no software package available that allows non-expert users to perform a full evaluation of such experiments. Here we present pyTFM, a tool to perform Traction Force Microscopy and Monolayer Stress Microscopy on cell patches and cell layers grown in a 2-dimensional environment. pyTFM was optimized for ease-of-use; it is open-source and well documented (hosted at https://pytfm.readthedocs.io/) including usage examples and explanations of the theoretical background. pyTFM can be used as a standalone Python package or as an add-on to the image annotation tool ClickPoints. In combination with the ClickPoints environment, pyTFM allows the user to set all necessary analysis parameters, select regions of interest, examine the input data and intermediary results, and calculate a wide range of parameters describing forces, stresses, and their distribution. In this work, we also thoroughly analyze the accuracy and performance of the Traction Force Microscopy and Monolayer Stress Microscopy algorithms of pyTFM using synthetic and experimental data from epithelial cell patches.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
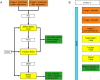
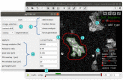


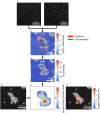
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

