Inhibitory Concentrations of Ciprofloxacin Induce an Adaptive Response Promoting the Intracellular Survival of Salmonella enterica Serovar Typhimurium
- PMID: 34154399
- PMCID: PMC8262899
- DOI: 10.1128/mBio.01093-21
Inhibitory Concentrations of Ciprofloxacin Induce an Adaptive Response Promoting the Intracellular Survival of Salmonella enterica Serovar Typhimurium
Abstract
Antimicrobial resistance (AMR) is a pressing global health crisis, which has been fueled by the sustained use of certain classes of antimicrobials, including fluoroquinolones. While the genetic mutations responsible for decreased fluoroquinolone (ciprofloxacin) susceptibility are known, the implications of ciprofloxacin exposure on bacterial growth, survival, and interactions with host cells are not well described. Aiming to understand the influence of inhibitory concentrations of ciprofloxacin in vitro, we subjected three clinical isolates of Salmonella enterica serovar Typhimurium to differing concentrations of ciprofloxacin, dependent on their MICs, and assessed the impact on bacterial growth, morphology, and transcription. We further investigated the differential morphology and transcription that occurred following ciprofloxacin exposure and measured the ability of ciprofloxacin-treated bacteria to invade and replicate in host cells. We found that ciprofloxacin-exposed S. Typhimurium is able to recover from inhibitory concentrations of ciprofloxacin and that the drug induces specific morphological and transcriptional signatures associated with the bacterial SOS response, DNA repair, and intracellular survival. In addition, ciprofloxacin-treated S. Typhimurium has increased capacity for intracellular replication in comparison to that of untreated organisms. These data suggest that S. Typhimurium undergoes an adaptive response under ciprofloxacin perturbation that promotes cellular survival, a consequence that may justify more measured use of ciprofloxacin for Salmonella infections. The combination of multiple experimental approaches provides new insights into the collateral effects that ciprofloxacin and other antimicrobials have on invasive bacterial pathogens. IMPORTANCE Antimicrobial resistance is a critical concern in global health. In particular, there is rising resistance to fluoroquinolones, such as ciprofloxacin, a first-line antimicrobial for many Gram-negative pathogens. We investigated the adaptive response of clinical isolates of Salmonella enterica serovar Typhimurium to ciprofloxacin, finding that the bacteria adapt in short timespans to high concentrations of ciprofloxacin in a way that promotes intracellular survival during early infection. Importantly, by studying three clinically relevant isolates, we were able to show that individual isolates respond differently to ciprofloxacin and that for each isolate, there was a heterogeneous response under ciprofloxacin treatment. The heterogeneity that arises from ciprofloxacin exposure may drive survival and proliferation of Salmonella during treatment and lead to drug resistance.
Keywords: AMR; Salmonella; antimicrobial agents; cellular morphology; ciprofloxacin; confocal microscopy; transcriptomics.
Figures

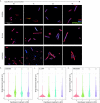
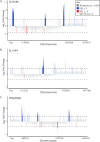
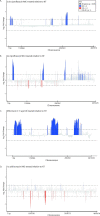

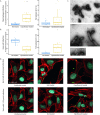
References
-
- Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, WHO Pathogens Priority List Working Group . 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. doi:10.1016/S1473-3099(17)30753-3. - DOI - PubMed
-
- O’Neill J. 2016. Tackling drug-resistant infections globally: final report and recommendations. Review on antimicrobial resistance, London, United Kingdom.
-
- Wohlkonig A, Chan PF, Fosberry AP, Homes P, Huang J, Kranz M, Leydon VR, Miles TJ, Pearson ND, Perera RL, Shillings AJ, Gwynn MN, Bax BD. 2010. Structural basis of quinolone inhibition of type IIA topoisomerases and target-mediated resistance. Nat Struct Mol Biol 17:1152–1153. doi:10.1038/nsmb.1892. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
