Single-Nucleus RNA Sequencing Identifies New Classes of Proximal Tubular Epithelial Cells in Kidney Fibrosis
- PMID: 34155061
- PMCID: PMC8722798
- DOI: 10.1681/ASN.2020081143
Single-Nucleus RNA Sequencing Identifies New Classes of Proximal Tubular Epithelial Cells in Kidney Fibrosis
Abstract
Background: Proximal tubular cells (PTCs) are the most abundant cell type in the kidney. PTCs are central to normal kidney function and to regeneration versus organ fibrosis following injury. This study used single-nucleus RNA sequencing (snRNAseq) to describe the phenotype of PTCs in renal fibrosis.
Methods: Kidneys were harvested from naïve mice and from mice with renal fibrosis induced by chronic aristolochic acid administration. Nuclei were isolated using Nuclei EZ Lysis buffer. Libraries were prepared on the 10× platform, and snRNAseq was completed using the Illumina NextSeq 550 System. Genome mapping was carried out with high-performance computing.
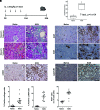
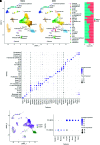
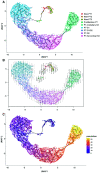
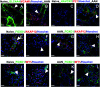
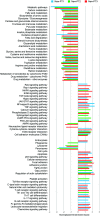
Results: A total of 23,885 nuclei were analyzed. PTCs were found in five abundant clusters, mapping to S1, S1-S2, S2, S2-cortical S3, and medullary S3 segments. Additional cell clusters ("new PTC clusters") were at low abundance in normal kidney and in increased number in kidneys undergoing regeneration/fibrosis following injury. These clusters exhibited clear molecular phenotypes, permitting labeling as proliferating, New-PT1, New-PT2, and (present only following injury) New-PT3. Each cluster exhibited a unique gene expression signature, including multiple genes previously associated with renal injury response and fibrosis progression. Comprehensive pathway analyses revealed metabolic reprogramming, enrichment of cellular communication and cell motility, and various immune activations in new PTC clusters. In ligand-receptor analysis, new PTC clusters promoted fibrotic signaling to fibroblasts and inflammatory activation to macrophages.
Conclusions: These data identify unrecognized PTC phenotype heterogeneity and reveal novel PTCs associated with kidney fibrosis.
Keywords: cell biology and structure; chronic kidney disease; epithelial; kidney tubule; mRNA; proximal tubule; renal epithelial cell; renal fibrosis; renal tubular epithelial cells; scRNA-seq.
Copyright © 2021 by the American Society of Nephrology.
Figures



References
-
- Meinild AK, Loo DD, Pajor AM, Zeuthen T, Wright EM: Water transport by the renal Na(+)-dicarboxylate cotransporter. Am J Physiol Renal Physiol 278: F777–F783, 2000 - PubMed
-
- Berg JMTJ, Stryer L: Biochemistry, 5th Ed., New York, W. H. Freeman, 2002
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

