Human intermediate progenitor diversity during cortical development
- PMID: 34155100
- PMCID: PMC8256045
- DOI: 10.1073/pnas.2019415118
Human intermediate progenitor diversity during cortical development
Abstract
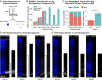
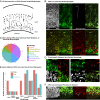
Studies of the spatiotemporal, transcriptomic, and morphological diversity of radial glia (RG) have spurred our current models of human corticogenesis. In the developing cortex, neural intermediate progenitor cells (nIPCs) are a neuron-producing transit-amplifying cell type born in the germinal zones of the cortex from RG. The potential diversity of the nIPC population, that produces a significant portion of excitatory cortical neurons, is understudied, particularly in the developing human brain. Here we explore the spatiotemporal, transcriptomic, and morphological variation that exists within the human nIPC population and provide a resource for future studies. We observe that the spatial distribution of nIPCs in the cortex changes abruptly around gestational week (GW) 19/20, marking a distinct shift in cellular distribution and organization during late neurogenesis. We also identify five transcriptomic subtypes, one of which appears at this spatiotemporal transition. Finally, we observe a diversity of nIPC morphologies that do not correlate with specific transcriptomic subtypes. These results provide an analysis of the spatiotemporal, transcriptional, and morphological diversity of nIPCs in developing brain tissue and provide an atlas of nIPC subtypes in the developing human cortex that can benchmark in vitro models of human development such as cerebral organoids and help inform future studies of how nIPCs contribute to cortical neurogenesis.
Keywords: cortex; development; human; neuronal; progenitor.
Conflict of interest statement
The authors declare no competing interest.
Figures





References
-
- Hansen D. V., Lui J. H., Parker P. R. L., Kriegstein A. R., Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 464, 554–561 (2010). - PubMed
-
- Hevner R. F., From radial glia to pyramidal-projection neuron: Transcription factor cascades in cerebral cortex development. Mol. Neurobiol. 33, 33–50 (2006). - PubMed
-
- Noctor S. C., Martínez-Cerdeño V., Ivic L., Kriegstein A. R., Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 7, 136–144 (2004). - PubMed

