Haplotype tagging reveals parallel formation of hybrid races in two butterfly species
- PMID: 34155138
- PMCID: PMC8237668
- DOI: 10.1073/pnas.2015005118
Haplotype tagging reveals parallel formation of hybrid races in two butterfly species
Abstract
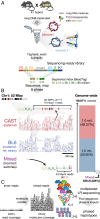
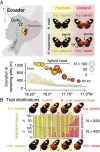
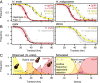
Genetic variation segregates as linked sets of variants or haplotypes. Haplotypes and linkage are central to genetics and underpin virtually all genetic and selection analysis. Yet, genomic data often omit haplotype information due to constraints in sequencing technologies. Here, we present "haplotagging," a simple, low-cost linked-read sequencing technique that allows sequencing of hundreds of individuals while retaining linkage information. We apply haplotagging to construct megabase-size haplotypes for over 600 individual butterflies (Heliconius erato and H. melpomene), which form overlapping hybrid zones across an elevational gradient in Ecuador. Haplotagging identifies loci controlling distinctive high- and lowland wing color patterns. Divergent haplotypes are found at the same major loci in both species, while chromosome rearrangements show no parallelism. Remarkably, in both species, the geographic clines for the major wing-pattern loci are displaced by 18 km, leading to the rise of a novel hybrid morph in the center of the hybrid zone. We propose that shared warning signaling (Müllerian mimicry) may couple the cline shifts seen in both species and facilitate the parallel coemergence of a novel hybrid morph in both comimetic species. Our results show the power of efficient haplotyping methods when combined with large-scale sequencing data from natural populations.
Keywords: butterfly; genomes; haplotypes; hybrid zone; population genetics.
Copyright © 2021 the Author(s). Published by PNAS.
Conflict of interest statement
Competing interest statement: M.K., A.D., and Y.F.C. declare competing financial interests in the form of patent and employment by the Max Planck Society. The European Research Council provides funding for the research but no other competing interests.
Figures


References
-
- Barton N. H., Keightley P. D., Understanding quantitative genetic variation. Nat. Rev. Genet. 3, 11–21 (2002). - PubMed
-
- Seehausen O., et al. ., Genomics and the origin of species. Nat. Rev. Genet. 15, 176–192(2014). - PubMed
-
- Sella G., Barton N. H., Thinking about the evolution of complex traits in the era of genome-wide association studies. Annu. Rev. Genomics Hum. Genet. 20, 461–493(2019). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

