ERRγ enhances cardiac maturation with T-tubule formation in human iPSC-derived cardiomyocytes
- PMID: 34155205
- PMCID: PMC8217550
- DOI: 10.1038/s41467-021-23816-3
ERRγ enhances cardiac maturation with T-tubule formation in human iPSC-derived cardiomyocytes
Abstract
One of the earliest maturation steps in cardiomyocytes (CMs) is the sarcomere protein isoform switch between TNNI1 and TNNI3 (fetal and neonatal/adult troponin I). Here, we generate human induced pluripotent stem cells (hiPSCs) carrying a TNNI1EmGFP and TNNI3mCherry double reporter to monitor and isolate mature sub-populations during cardiac differentiation. Extensive drug screening identifies two compounds, an estrogen-related receptor gamma (ERRγ) agonist and an S-phase kinase-associated protein 2 inhibitor, that enhances cardiac maturation and a significant change to TNNI3 expression. Expression, morphological, functional, and molecular analyses indicate that hiPSC-CMs treated with the ERRγ agonist show a larger cell size, longer sarcomere length, the presence of transverse tubules, and enhanced metabolic function and contractile and electrical properties. Here, we show that ERRγ-treated hiPSC-CMs have a mature cellular property consistent with neonatal CMs and are useful for disease modeling and regenerative medicine.
Conflict of interest statement
Y.Y. received research expense from Takeda Pharmaceutical Company, Ltd, and K.D., S.K., S.C.N., S.I, T.M., T.A., N.T., K.I., T.N. are employees of Takeda Pharmaceutical Company, Ltd. K.M., S.K., and Y.Y. are the inventors of the patent application (WO2019/189554). The other authors declare no competing interests.
Figures
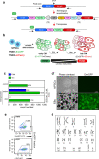
 blue bars; VIC probe,
blue bars; VIC probe,  green bars) of TNNI1EmGFP knock-in clones before and after treatment with piggyBac transposase. d Phase-contrast and EmGFP fluorescence images of hiPSCs (#4-12-20) and day-14 EBs. Scale bars: 500 μm. e Flow cytometric analysis and sorting on day 14 of the differentiation for EmGFP+ cells (top) and the EmGFP+-derived cTNT+ cell fraction (bottom). f Karyotype of the generated TNNI1-EmGFP knockin clone #4-12-20.
green bars) of TNNI1EmGFP knock-in clones before and after treatment with piggyBac transposase. d Phase-contrast and EmGFP fluorescence images of hiPSCs (#4-12-20) and day-14 EBs. Scale bars: 500 μm. e Flow cytometric analysis and sorting on day 14 of the differentiation for EmGFP+ cells (top) and the EmGFP+-derived cTNT+ cell fraction (bottom). f Karyotype of the generated TNNI1-EmGFP knockin clone #4-12-20.

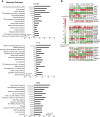
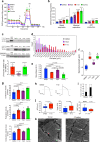
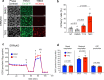
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

