Insights into how environment shapes post-mortem RNA transcription in mouse brain
- PMID: 34155272
- PMCID: PMC8217559
- DOI: 10.1038/s41598-021-92268-y
Insights into how environment shapes post-mortem RNA transcription in mouse brain
Abstract
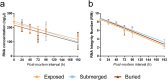
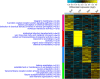
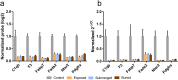
Most biological features that occur on the body after death were already deciphered by traditional medicine. However, the molecular mechanisms triggered in the cellular microenvironment are not fully comprehended yet. Previous studies reported gene expression alterations in the post-mortem condition, but little is known about how the environment could influence RNA degradation and transcriptional regulation. In this work, we analysed the transcriptome of mouse brain after death under three concealment simulations (air exposed, buried, and submerged). Our analyses identified 2,103 genes differentially expressed in all tested groups 48 h after death. Moreover, we identified 111 commonly upregulated and 497 commonly downregulated genes in mice from the concealment simulations. The gene functions shared by the individuals from the tested environments were associated with RNA homeostasis, inflammation, developmental processes, cell communication, cell proliferation, and lipid metabolism. Regarding the altered biological processes, we identified that the macroautophagy process was enriched in the upregulated genes and lipid metabolism was enriched in the downregulated genes. On the other hand, we also described a list of biomarkers associated with the submerged and buried groups, indicating that these environments can influence the post-mortem RNA abundance in its particular way.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

