Compositional biases in RNA viruses: Causes, consequences and applications
- PMID: 34155814
- PMCID: PMC8420353
- DOI: 10.1002/wrna.1679
Compositional biases in RNA viruses: Causes, consequences and applications
Abstract
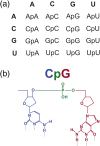
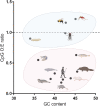
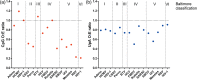
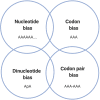
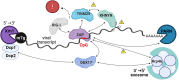
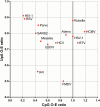
If each of the four nucleotides were represented equally in the genomes of viruses and the hosts they infect, each base would occur at a frequency of 25%. However, this is not observed in nature. Similarly, the order of nucleotides is not random (e.g., in the human genome, guanine follows cytosine at a frequency of ~0.0125, or a quarter the number of times predicted by random representation). Codon usage and codon order are also nonrandom. Furthermore, nucleotide and codon biases vary between species. Such biases have various drivers, including cellular proteins that recognize specific patterns in nucleic acids, that once triggered, induce mutations or invoke intrinsic or innate immune responses. In this review we examine the types of compositional biases identified in viral genomes and current understanding of the evolutionary mechanisms underpinning these trends. Finally, we consider the potential for large scale synonymous recoding strategies to engineer RNA virus vaccines, including those with pandemic potential, such as influenza A virus and Severe Acute Respiratory Syndrome Coronavirus Virus 2. This article is categorized under: RNA in Disease and Development > RNA in Disease RNA Evolution and Genomics > Computational Analyses of RNA RNA Interactions with Proteins and Other Molecules > Protein-RNA Recognition.
Keywords: dinucleotides; mutation bias; selection bias; viral genome composition.
© 2021 The Authors. WIREs RNA published by Wiley Periodicals LLC.
Conflict of interest statement
The authors have declared no conflicts of interest for this article.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

