Adipsin promotes bone marrow adiposity by priming mesenchymal stem cells
- PMID: 34155972
- PMCID: PMC8219379
- DOI: 10.7554/eLife.69209
Adipsin promotes bone marrow adiposity by priming mesenchymal stem cells
Abstract
Background: Marrow adipose tissue (MAT) has been shown to be vital for regulating metabolism and maintaining skeletal homeostasis in the bone marrow (BM) niche. As a reflection of BM remodeling, MAT is highly responsive to nutrient fluctuations, hormonal changes, and metabolic disturbances such as obesity and diabetes mellitus. Expansion of MAT has also been strongly associated with bone loss in mice and humans. However, the regulation of BM plasticity remains poorly understood, as does the mechanism that links changes in marrow adiposity with bone remodeling.
Methods: We studied deletion of Adipsin, and its downstream effector, C3, in C57BL/6 mice as well as the bone-protected PPARγ constitutive deacetylation 2KR mice to assess BM plasticity. The mice were challenged with thiazolidinedione treatment, calorie restriction, or aging to induce bone loss and MAT expansion. Analysis of bone mineral density and marrow adiposity was performed using a μCT scanner and by RNA analysis to assess adipocyte and osteoblast markers. For in vitro studies, primary bone marrow stromal cells were isolated and subjected to osteoblastogenic or adipogenic differentiation or chemical treatment followed by morphological and molecular analyses. Clinical data was obtained from samples of a previous clinical trial of fasting and high-calorie diet in healthy human volunteers.
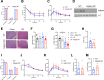
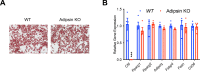
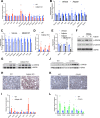
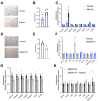
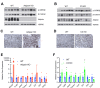
Results: We show that Adipsin is the most upregulated adipokine during MAT expansion in mice and humans in a PPARγ acetylation-dependent manner. Genetic ablation of Adipsin in mice specifically inhibited MAT expansion but not peripheral adipose depots, and improved bone mass during calorie restriction, thiazolidinedione treatment, and aging. These effects were mediated through its downstream effector, complement component C3, to prime common progenitor cells toward adipogenesis rather than osteoblastogenesis through inhibiting Wnt/β-catenin signaling.
Conclusions: Adipsin promotes new adipocyte formation and affects skeletal remodeling in the BM niche. Our study reveals a novel mechanism whereby the BM sustains its own plasticity through paracrine and endocrine actions of a unique adipokine.
Funding: This work was supported by the National Institutes of Health T32DK007328 (NA), F31DK124926 (NA), R01DK121140 (JCL), R01AR068970 (BZ), R01AR071463 (BZ), R01DK112943 (LQ), R24DK092759 (CJR), and P01HL087123 (LQ).
Keywords: PPARgamma; adipokine; adipsin; bone marrow adiposity; complement; medicine; mouse.
© 2021, Aaron et al.
Conflict of interest statement
NA, MK, QZ, QL, SC, JY, LL, LY, LW, YH, LF, HH, LD, JL, WW, BZ, EG, LS, CR, LQ No competing interests declared
Figures












References
-
- Bloom AC, Collins FL, Van't Hof RJ, Ryan ES, Jones E, Hughes TR, Morgan BP, Erlandsson M, Bokarewa M, Aeschlimann D, Evans BAJ, Williams AS. Deletion of the membrane complement inhibitor CD59a drives age and gender-dependent alterations to bone phenotype in mice. Bone. 2016;84:253–261. doi: 10.1016/j.bone.2015.12.014. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

