Untargeted Metabolomics Reveals Species-Specific Metabolite Production and Shared Nutrient Consumption by Pseudomonas aeruginosa and Staphylococcus aureus
- PMID: 34156287
- PMCID: PMC8269234
- DOI: 10.1128/mSystems.00480-21
Untargeted Metabolomics Reveals Species-Specific Metabolite Production and Shared Nutrient Consumption by Pseudomonas aeruginosa and Staphylococcus aureus
Abstract
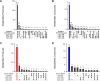
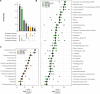
While bacterial metabolism is known to impact antibiotic efficacy and virulence, the metabolic capacities of individual microbes in cystic fibrosis lung infections are difficult to disentangle from sputum samples. Here, we show that untargeted metabolomic profiling of supernatants of multiple strains of Pseudomonas aeruginosa and Staphylococcus aureus grown in monoculture in synthetic cystic fibrosis media (SCFM) reveals distinct species-specific metabolic signatures despite intraspecies metabolic variability. We identify a set of 15 metabolites that were significantly consumed by both P. aeruginosa and S. aureus, suggesting that nutrient competition has the potential to impact community dynamics even in the absence of other pathogen-pathogen interactions. Finally, metabolites that were uniquely produced by one species or the other were identified. Specifically, the virulence factor precursor anthranilic acid, as well as the quinoline 2,4-quinolinediol (DHQ), were robustly produced across all tested strains of P. aeruginosa. Through the direct comparison of the extracellular metabolism of P. aeruginosa and S. aureus in a physiologically relevant environment, this work provides insight toward the potential for metabolic interactions in vivo and supports the development of species-specific diagnostic markers of infection. IMPORTANCE Interactions between P. aeruginosa and S. aureus can impact pathogenicity and antimicrobial efficacy. In this study, we aim to better understand the potential for metabolic interactions between P. aeruginosa and S. aureus in an environment resembling the cystic fibrosis lung. We find that S. aureus and P. aeruginosa consume many of the same nutrients, suggesting that metabolic competition may play an important role in community dynamics during coinfection. We further identify metabolites uniquely produced by either organism with the potential to be developed into species-specific biomarkers of infection in the cystic fibrosis lung.
Keywords: LC-MS; Pseudomonas aeruginosa; Staphylococcus aureus; metabolism.
Figures



References
-
- Cystic Fibrosis Foundation. 2018. 2018 Patient Registry Annual Data Report. Cystic Fibrosis Foundation, Bethesda, Maryland.
-
- Limoli DH, Yang J, Khansaheb MK, Helfman B, Peng L, Stecenko AA, Goldberg JB. 2016. Staphylococcus aureus and Pseudomonas aeruginosa co-infection is associated with cystic fibrosis-related diabetes and poor clinical outcomes. 6. Eur J Clin Microbiol Infect Dis 35:947–953. doi:10.1007/s10096-016-2621-0. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

