Long-Term Metabolomics Reference Material
- PMID: 34156835
- PMCID: PMC8996483
- DOI: 10.1021/acs.analchem.1c01294
Long-Term Metabolomics Reference Material
Abstract
The use of quality control samples in metabolomics ensures data quality, reproducibility, and comparability between studies, analytical platforms, and laboratories. Long-term, stable, and sustainable reference materials (RMs) are a critical component of the quality assurance/quality control (QA/QC) system; however, the limited selection of currently available matrix-matched RMs reduces their applicability for widespread use. To produce an RM in any context, for any matrix that is robust to changes over the course of time, we developed iterative batch averaging method (IBAT). To illustrate this method, we generated 11 independently grown Escherichia coli batches and made an RM over the course of 10 IBAT iterations. We measured the variance of these materials by nuclear magnetic resonance (NMR) and showed that IBAT produces a stable and sustainable RM over time. This E. coli RM was then used as a food source to produce a Caenorhabditis elegans RM for a metabolomics experiment. The metabolite extraction of this material, alongside 41 independently grown individual C. elegans samples of the same genotype, allowed us to estimate the proportion of sample variation in preanalytical steps. From the NMR data, we found that 40% of the metabolite variance is due to the metabolite extraction process and analysis and 60% is due to sample-to-sample variance. The availability of RMs in untargeted metabolomics is one of the predominant needs of the metabolomics community that reach beyond quality control practices. IBAT addresses this need by facilitating the production of biologically relevant RMs and increasing their widespread use.
Figures
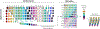
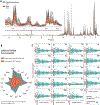

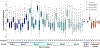
References
-
- Cochran WG; Cox GM Experimental Designs, 2nd ed., 1957.
-
- Dunn WB; Broadhurst DI; Edison A; Guillou C; Viant MR; Bearden DW; Beger RD Metabolomics 2017, 13, No. 50.
-
- Paulovich AG; Billheimer D; Ham AJ; Vega-Montoto L; Rudnick PA; Tabb DL; Wang P; Blackman RK; Bunk DM; Cardasis HL; Clauser KR; Kinsinger CR; Schilling B; Tegeler TJ; Variyath AM; Wang M; Whiteaker JR; Zimmerman LJ; Fenyo D; Carr SA; Fisher SJ; Gibson BW; Mesri M; Neubert TA; Regnier FE; Rodriguez H; Spiegelman C; Stein SE; Tempst P; Liebler DC Mol. Cell. Proteomics 2010, 9, 242–254. - PMC - PubMed
-
- Phinney KW; Ballihaut G; Bedner M; Benford BS; Camara JE; Christopher SJ; Davis WC; Dodder NG; Eppe G; Lang BE; Long SE; Lowenthal MS; McGaw EA; Murphy KE; Nelson BC; Prendergast JL; Reiner JL; Rimmer CA; Sander LC; Schantz MM; Sharpless KE; Sniegoski LT; Tai SS; Thomas JB; Vetter TW; Welch MJ; Wise SA; Wood LJ; Guthrie WF; Hagwood CR; Leigh SD; Yen JH; Zhang NF; Chaudhary-Webb M; Chen H; Fazili Z; LaVoie DJ; McCoy LF; Momin SS; Paladugula N; Pendergrast EC; Pfeiffer CM; Powers CD; Rabinowitz D; Rybak ME; Schleicher RL; Toombs BM; Xu M; Zhang M; Castle AL Anal. Chem 2013, 85, 11732–11738. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

