Endothelial STING controls T cell transmigration in an IFNI-dependent manner
- PMID: 34156982
- PMCID: PMC8410041
- DOI: 10.1172/jci.insight.149346
Endothelial STING controls T cell transmigration in an IFNI-dependent manner
Abstract
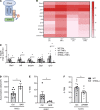
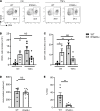
The stimulator of IFN genes (STING) protein senses cyclic dinucleotides released in response to double-stranded DNA and functions as an adaptor molecule for type I IFN (IFNI) signaling by activating IFNI-stimulated genes (ISG). We found impaired T cell infiltration into the peritoneum in response to TNF-α in global and EC-specific STING-/- mice and discovered that T cell transendothelial migration (TEM) across mouse and human endothelial cells (EC) deficient in STING was strikingly reduced compared with control EC, whereas T cell adhesion was not impaired. STING-/- T cells showed no defect in TEM or adhesion to EC, or immobilized endothelial cell-expressed molecules ICAM1 and VCAM1, compared with WT T cells. Mechanistically, CXCL10, an ISG and a chemoattractant for T cells, was dramatically reduced in TNF-α-stimulated STING-/- EC, and genetic loss or pharmacologic antagonisms of IFNI receptor (IFNAR) pathway reduced T cell TEM. Our data demonstrate a central role for EC-STING during T cell TEM that is dependent on the ISG CXCL10 and on IFNI/IFNAR signaling.
Keywords: Cell migration/adhesion; Endothelial cells; Inflammation; Vascular Biology.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

