Effect of clinical isolate or cleavage site mutations in the SARS-CoV-2 spike protein on protein stability, cleavage, and cell-cell fusion
- PMID: 34157282
- PMCID: PMC8214756
- DOI: 10.1016/j.jbc.2021.100902
Effect of clinical isolate or cleavage site mutations in the SARS-CoV-2 spike protein on protein stability, cleavage, and cell-cell fusion
Abstract
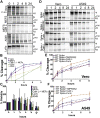
The trimeric severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein (S) is the sole viral protein responsible for both viral binding to a host cell and the membrane fusion event needed for cell entry. In addition to facilitating fusion needed for viral entry, S can also drive cell-cell fusion, a pathogenic effect observed in the lungs of SARS-CoV-2-infected patients. While several studies have investigated S requirements involved in viral particle entry, examination of S stability and factors involved in S cell-cell fusion remain limited. A furin cleavage site at the border between the S1 and S2 subunits (S1/S2) has been identified, along with putative cathepsin L and transmembrane serine protease 2 cleavage sites within S2. We demonstrate that S must be processed at the S1/S2 border in order to mediate cell-cell fusion and that mutations at potential cleavage sites within the S2 subunit alter S processing at the S1/S2 border, thus preventing cell-cell fusion. We also identify residues within the internal fusion peptide and the cytoplasmic tail that modulate S-mediated cell-cell fusion. In addition, we examined S stability and protein cleavage kinetics in a variety of mammalian cell lines, including a bat cell line related to the likely reservoir species for SARS-CoV-2, and provide evidence that proteolytic processing alters the stability of the S trimer. This work therefore offers insight into S stability, proteolytic processing, and factors that mediate S cell-cell fusion, all of which help give a more comprehensive understanding of this high-profile therapeutic target.
Keywords: COVID-19; SARS-CoV-2; coronavirus; fusion protein; membrane fusion; viral protein; virology; virus entry.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures




Update of
-
Effect of mutations in the SARS-CoV-2 spike protein on protein stability, cleavage, and cell-cell fusion function.bioRxiv [Preprint]. 2021 Jan 25:2021.01.24.428007. doi: 10.1101/2021.01.24.428007. bioRxiv. 2021. Update in: J Biol Chem. 2021 Jul;297(1):100902. doi: 10.1016/j.jbc.2021.100902. PMID: 33532777 Free PMC article. Updated. Preprint.
References
-
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

