DotMotif: an open-source tool for connectome subgraph isomorphism search and graph queries
- PMID: 34158519
- PMCID: PMC8219732
- DOI: 10.1038/s41598-021-91025-5
DotMotif: an open-source tool for connectome subgraph isomorphism search and graph queries
Abstract
Recent advances in neuroscience have enabled the exploration of brain structure at the level of individual synaptic connections. These connectomics datasets continue to grow in size and complexity; methods to search for and identify interesting graph patterns offer a promising approach to quickly reduce data dimensionality and enable discovery. These graphs are often too large to be analyzed manually, presenting significant barriers to searching for structure and testing hypotheses. We combine graph database and analysis libraries with an easy-to-use neuroscience grammar suitable for rapidly constructing queries and searching for subgraphs and patterns of interest. Our approach abstracts many of the computer science and graph theory challenges associated with nanoscale brain network analysis and allows scientists to quickly conduct research at scale. We demonstrate the utility of these tools by searching for motifs on simulated data and real public connectomics datasets, and we share simple and complex structures relevant to the neuroscience community. We contextualize our findings and provide case studies and software to motivate future neuroscience exploration.
Conflict of interest statement
The authors declare no competing interests.
Figures
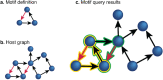

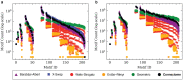
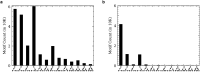
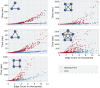



References
-
- Scheffer LK. Graph properties of the adult drosophila central brain. biorXiv. 2020 doi: 10.1101/2020.05.18.102061. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

