The first genotype II African swine fever virus isolated in Africa provides insight into the current Eurasian pandemic
- PMID: 34158551
- PMCID: PMC8219699
- DOI: 10.1038/s41598-021-92593-2
The first genotype II African swine fever virus isolated in Africa provides insight into the current Eurasian pandemic
Abstract
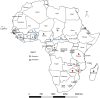
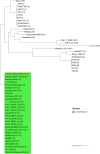
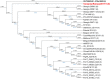
African swine fever (ASF) caused by the African swine fever virus (ASFV) is ranked by OIE as the most important source of mortality in domestic pigs globally and is indigenous to African wild suids and soft ticks. Despite two ASFV genotypes causing economically devastating epidemics outside the continent since 1961, there have been no genome-level analyses of virus evolution in Africa. The virus was recently transported from south-eastern Africa to Georgia in 2007 and has subsequently spread to Russia, eastern Europe, China, and south-east Asia with devastating socioeconomic consequences. To date, two of the 24 currently described ASFV genotypes defined by sequencing of the p72 gene, namely genotype I and II, have been reported outside Africa, with genotype II being responsible for the ongoing pig pandemic. Multiple complete genotype II genome sequences have been reported from European, Russian and Chinese virus isolates but no complete genome sequences have yet been reported from Africa. We report herein the complete genome of a Tanzanian genotype II isolate, Tanzania/Rukwa/2017/1, collected in 2017 and determined using an Illumina short read strategy. The Tanzania/Rukwa/2017/1 sequence is 183,186 bp in length (in a single contig) and contains 188 open reading frames. Considering only un-gapped sites in the pairwise alignments, the new sequence has 99.961% identity with the updated Georgia 2007/1 reference isolate (FR682468.2), 99.960% identity with Polish isolate Pol16_29413_o23 (MG939586) and 99.957% identity with Chinese isolate ASFV-wbBS01 (MK645909.1). This represents 73 single nucleotide polymorphisms (SNPs) relative to the Polish isolate and 78 SNPs with the Chinese genome. Phylogenetic analysis indicated that Tanzania/Rukwa/2017/1 clusters most closely with Georgia 2007/1. The majority of the differences between Tanzania/Rukwa/2017/1 and Georgia 2007/1 genotype II genomes are insertions/deletions (indels) as is typical for ASFV. The indels included differences in the length and copy number of the terminal multicopy gene families, MGF 360 and 110. The Rukwa2017/1 sequence is the first complete genotype II genome from a precisely mapped locality in Africa, since the exact origin of Georgia2007/1 is unknown. It therefore provides baseline information for future analyses of the diversity and phylogeography of this globally important genetic sub-group of ASF viruses.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- OIE-Listed diseases 2019: OIE—World Organisation for Animal Health. http://www.oie.int/en/animal-health-in-the-world/oie-listed-diseases-2019/.
-
- Eustace Montgomery R. On a form of swine fever occurring in British East Africa (Kenya Colony) J. Comp. Pathol. Ther. 1921;34:159–191. doi: 10.1016/S0368-1742(21)80031-4. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

