Growth faltering regardless of chronic diarrhea is associated with mucosal immune dysfunction and microbial dysbiosis in the gut lumen
- PMID: 34158595
- PMCID: PMC8379072
- DOI: 10.1038/s41385-021-00418-2
Growth faltering regardless of chronic diarrhea is associated with mucosal immune dysfunction and microbial dysbiosis in the gut lumen
Abstract
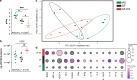
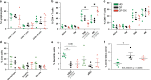
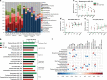
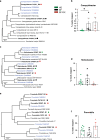
Despite the impact of childhood diarrhea on morbidity and mortality, our understanding of its sequelae has been significantly hampered by the lack of studies that examine samples across the entire intestinal tract. Infant rhesus macaques are naturally susceptible to human enteric pathogens and recapitulate the hallmarks of diarrheal disease such as intestinal inflammation and growth faltering. Here, we examined intestinal biopsies, lamina propria leukocytes, luminal contents, and fecal samples from healthy infants and those experiencing growth faltering with distant acute or chronic active diarrhea. We show that growth faltering in the presence or absence of active diarrhea is associated with a heightened systemic and mucosal pro-inflammatory state centered in the colon. Moreover, polyclonal stimulation of colonic lamina propria leukocytes resulted in a dampened cytokine response, indicative of immune exhaustion. We also detected a functional and taxonomic shift in the luminal microbiome across multiple gut sites including the migration of Streptococcus and Prevotella species between the small and large intestine, suggesting a decompartmentalization of gut microbial communities. Our studies provide valuable insight into the outcomes of diarrheal diseases and growth faltering not attainable in humans and lays the groundwork to test interventions in a controlled and reproducible setting.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Dietary Gluten-Induced Gut Dysbiosis Is Accompanied by Selective Upregulation of microRNAs with Intestinal Tight Junction and Bacteria-Binding Motifs in Rhesus Macaque Model of Celiac Disease.Nutrients. 2016 Oct 28;8(11):684. doi: 10.3390/nu8110684. Nutrients. 2016. PMID: 27801835 Free PMC article.
-
S100A8 and S100A9 Are Important for Postnatal Development of Gut Microbiota and Immune System in Mice and Infants.Gastroenterology. 2020 Dec;159(6):2130-2145.e5. doi: 10.1053/j.gastro.2020.08.019. Epub 2020 Aug 15. Gastroenterology. 2020. PMID: 32805279
-
Antigen-Specific Mucosal Immunity Regulates Development of Intestinal Bacteria-Mediated Diseases.Gastroenterology. 2019 Dec;157(6):1530-1543.e4. doi: 10.1053/j.gastro.2019.08.021. Epub 2019 Aug 21. Gastroenterology. 2019. PMID: 31445037
-
Gut microbiome in primary sclerosing cholangitis: A review.World J Gastroenterol. 2020 Jun 7;26(21):2768-2780. doi: 10.3748/wjg.v26.i21.2768. World J Gastroenterol. 2020. PMID: 32550753 Free PMC article. Review.
-
Development, validation and implementation of an in vitro model for the study of metabolic and immune function in normal and inflamed human colonic epithelium.Dan Med J. 2015 Jan;62(1):B4973. Dan Med J. 2015. PMID: 25557335 Review.
Cited by
-
Phenotypic and Functional Alterations in Peripheral Blood Mononuclear Cell-Derived Microglia in a Primate Model of Chronic Alcohol Consumption.bioRxiv [Preprint]. 2025 Feb 6:2025.02.05.636708. doi: 10.1101/2025.02.05.636708. bioRxiv. 2025. Update in: Brain Behav Immun. 2025 Jul 26;129:874-889. doi: 10.1016/j.bbi.2025.07.022. PMID: 39975066 Free PMC article. Updated. Preprint.
-
Effect of a Homestead Food Production and Food Hygiene Intervention on Biomarkers of Environmental Enteric Dysfunction in Children Younger Than 24 Months in Rural Bangladesh: A Cluster-Randomized Controlled Trial.Am J Trop Med Hyg. 2023 Oct 2;109(5):1166-1176. doi: 10.4269/ajtmh.23-0153. Print 2023 Nov 1. Am J Trop Med Hyg. 2023. PMID: 37783459 Free PMC article. Clinical Trial.
-
Gut microbiota bridges the iron homeostasis and host health.Sci China Life Sci. 2023 Sep;66(9):1952-1975. doi: 10.1007/s11427-022-2302-5. Epub 2023 Jul 27. Sci China Life Sci. 2023. PMID: 37515687 Review.
-
Water, sanitation, hygiene practices, health and nutritional status among children before and during the COVID-19 pandemic: longitudinal evidence from remote areas of Dailekh and Achham districts in Nepal.BMC Public Health. 2022 Nov 7;22(1):2035. doi: 10.1186/s12889-022-14346-8. BMC Public Health. 2022. PMID: 36344970 Free PMC article.
-
Coated Zinc Oxide Improves Growth Performance of Weaned Piglets via Gut Microbiota.Front Nutr. 2022 Feb 25;9:819722. doi: 10.3389/fnut.2022.819722. eCollection 2022. Front Nutr. 2022. PMID: 35284437 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

