Histone acylations and chromatin dynamics: concepts, challenges, and links to metabolism
- PMID: 34159701
- PMCID: PMC8406397
- DOI: 10.15252/embr.202152774
Histone acylations and chromatin dynamics: concepts, challenges, and links to metabolism
Abstract
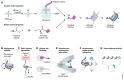
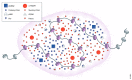
In eukaryotic cells, DNA is tightly packed with the help of histone proteins into chromatin. Chromatin architecture can be modified by various post-translational modifications of histone proteins. For almost 60 years now, studies on histone lysine acetylation have unraveled the contribution of this acylation to an open chromatin state with increased DNA accessibility, permissive for gene expression. Additional complexity emerged from the discovery of other types of histone lysine acylations. The acyl group donors are products of cellular metabolism, and distinct histone acylations can link the metabolic state of a cell with chromatin architecture and contribute to cellular adaptation through changes in gene expression. Currently, various technical challenges limit our full understanding of the actual impact of most histone acylations on chromatin dynamics and of their biological relevance. In this review, we summarize the state of the art and provide an overview of approaches to overcome these challenges. We further discuss the concept of subnuclear metabolic niches that could regulate local CoA availability and thus couple cellular metabolisms with the epigenome.
Keywords: acylation; chromatin; histones; metabolism; microdomains.
© 2021 The Authors. Published under the terms of the CC BY NC ND 4.0 license.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Allis CD, Jenuwein T (2016) The molecular hallmarks of epigenetic control. Nat Rev Genet 17: 487–500 - PubMed
-
- Bao X, Liu Z, Zhang W, Gladysz K, Fung YME, Tian G, Xiong Y, Wong JWH, Yuen KWY, Li XD (2019) Glutarylation of histone H4 lysine 91 regulates chromatin dynamics. Mol Cell 76: 660–675.e9 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

