Mining and unearthing hidden biosynthetic potential
- PMID: 34162873
- PMCID: PMC8222398
- DOI: 10.1038/s41467-021-24133-5
Mining and unearthing hidden biosynthetic potential
Abstract
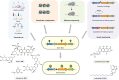
Genetically encoded small molecules (secondary metabolites) play eminent roles in ecological interactions, as pathogenicity factors and as drug leads. Yet, these chemical mediators often evade detection, and the discovery of novel entities is hampered by low production and high rediscovery rates. These limitations may be addressed by genome mining for biosynthetic gene clusters, thereby unveiling cryptic metabolic potential. The development of sophisticated data mining methods and genetic and analytical tools has enabled the discovery of an impressive array of previously overlooked natural products. This review shows the newest developments in the field, highlighting compound discovery from unconventional sources and microbiomes.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

