Human cytomegalovirus-induced host protein citrullination is crucial for viral replication
- PMID: 34162877
- PMCID: PMC8222335
- DOI: 10.1038/s41467-021-24178-6
Human cytomegalovirus-induced host protein citrullination is crucial for viral replication
Abstract
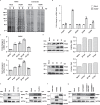
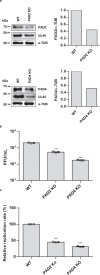
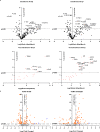
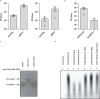
Citrullination is the conversion of arginine-to-citrulline by protein arginine deiminases (PADs), whose dysregulation is implicated in the pathogenesis of various types of cancers and autoimmune diseases. Consistent with the ability of human cytomegalovirus (HCMV) to induce post-translational modifications of cellular proteins to gain a survival advantage, we show that HCMV infection of primary human fibroblasts triggers PAD-mediated citrullination of several host proteins, and that this activity promotes viral fitness. Citrullinome analysis reveals significant changes in deimination levels of both cellular and viral proteins, with interferon (IFN)-inducible protein IFIT1 being among the most heavily deiminated one. As genetic depletion of IFIT1 strongly enhances HCMV growth, and in vitro IFIT1 citrullination impairs its ability to bind to 5'-ppp-RNA, we propose that viral-induced IFIT1 citrullination is a mechanism of HCMV evasion from host antiviral resistance. Overall, our findings point to a crucial role of citrullination in subverting cellular responses to viral infection.
Conflict of interest statement
The authors declare the following competing financial interest(s): P.R.T. founded Padlock Therapeutics and is entitled to payments from Bristol Myers Squibb if certain milestones are met. P.R.T. is a consultant for Celgene and Disarm Therapeutics. Italian Patent No. 102011596547852 issued in October 20, 2019 (“PAD2 PER USO NELLA PREVENZIONE E/O TRATTAMENTO O DIAGNOSI DI INFEZIONI DA VIRUS DELLA FAMIGLIA HERPESVIRIDAE”).
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

