ATRX promotes heterochromatin formation to protect cells from G-quadruplex DNA-mediated stress
- PMID: 34162889
- PMCID: PMC8222256
- DOI: 10.1038/s41467-021-24206-5
ATRX promotes heterochromatin formation to protect cells from G-quadruplex DNA-mediated stress
Abstract
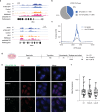
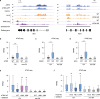
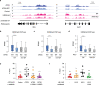
ATRX is a tumor suppressor that has been associated with protection from DNA replication stress, purportedly through resolution of difficult-to-replicate G-quadruplex (G4) DNA structures. While several studies demonstrate that loss of ATRX sensitizes cells to chemical stabilizers of G4 structures, the molecular function of ATRX at G4 regions during replication remains unknown. Here, we demonstrate that ATRX associates with a number of the MCM replication complex subunits and that loss of ATRX leads to G4 structure accumulation at newly synthesized DNA. We show that both the helicase domain of ATRX and its H3.3 chaperone function are required to protect cells from G4-induced replicative stress. Furthermore, these activities are upstream of heterochromatin formation mediated by the histone methyltransferase, ESET, which is the critical molecular event that protects cells from G4-mediated stress. In support, tumors carrying mutations in either ATRX or ESET show increased mutation burden at G4-enriched DNA sequences. Overall, our study provides new insights into mechanisms by which ATRX promotes genome stability with important implications for understanding impacts of its loss on human disease.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

