Ultrasensitive single-cell proteomics workflow identifies >1000 protein groups per mammalian cell
- PMID: 34163866
- PMCID: PMC8178986
- DOI: 10.1039/d0sc03636f
Ultrasensitive single-cell proteomics workflow identifies >1000 protein groups per mammalian cell
Abstract
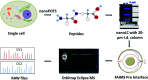
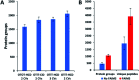
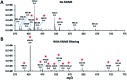
We report on the combination of nanodroplet sample preparation, ultra-low-flow nanoLC, high-field asymmetric ion mobility spectrometry (FAIMS), and the latest-generation Orbitrap Eclipse Tribrid mass spectrometer for greatly improved single-cell proteome profiling. FAIMS effectively filtered out singly charged ions for more effective MS analysis of multiply charged peptides, resulting in an average of 1056 protein groups identified from single HeLa cells without MS1-level feature matching. This is 2.3 times more identifications than without FAIMS and a far greater level of proteome coverage for single mammalian cells than has been previously reported for a label-free study. Differential analysis of single microdissected motor neurons and interneurons from human spinal tissue indicated a similar level of proteome coverage, and the two subpopulations of cells were readily differentiated based on single-cell label-free quantification.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

