Probing conformational hotspots for the recognition and intervention of protein complexes by lysine reactivity profiling
- PMID: 34163908
- PMCID: PMC8179027
- DOI: 10.1039/d0sc05330a
Probing conformational hotspots for the recognition and intervention of protein complexes by lysine reactivity profiling
Abstract
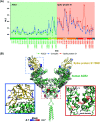
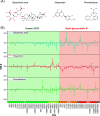
Probing the conformational and functional hotspot sites within aqueous native protein complexes is still a challenging task. Herein, a mass spectrometry (MS)-based two-step isotope labeling-lysine reactivity profiling (TILLRP) strategy is developed to quantify the reactivities of lysine residues and probe the molecular details of protein-protein interactions as well as evaluate the conformational interventions by small-molecule active compounds. The hotspot lysine sites that are crucial to the SARS-CoV-2 S1-ACE2 combination could be successfully probed, such as S1 Lys417 and Lys444. Significant alteration of the reactivities of lysine residues at the interaction interface of S1-RBD Lys386-Lys462 was observed during the formation of complexes, which might be utilized as indicators for investigating the S1-ACE2 dynamic recognition and intervention at the molecular level in high throughput.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures

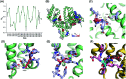
Similar articles
-
Quantitative Lysine Reactivity Profiling Reveals Conformational Inhibition Dynamics and Potency of Aurora A Kinase Inhibitors.Anal Chem. 2019 Oct 15;91(20):13222-13229. doi: 10.1021/acs.analchem.9b03647. Epub 2019 Sep 26. Anal Chem. 2019. PMID: 31525957
-
Probing the Lysine Proximal Microenvironments within Membrane Protein Complexes by Active Dimethyl Labeling and Mass Spectrometry.Anal Chem. 2016 Dec 20;88(24):12060-12065. doi: 10.1021/acs.analchem.6b02502. Epub 2016 Dec 6. Anal Chem. 2016. PMID: 28193046
-
Structural characterization of protein-material interfacial interactions using lysine reactivity profiling-mass spectrometry.Nat Protoc. 2023 Aug;18(8):2600-2623. doi: 10.1038/s41596-023-00849-0. Epub 2023 Jul 17. Nat Protoc. 2023. PMID: 37460632 Review.
-
Change of reactivity of lysine residues upon actin polymerization.Biochemistry. 1981 Sep 29;20(20):5914-9. doi: 10.1021/bi00523a040. Biochemistry. 1981. PMID: 6794619
-
Chemical cross-linking and mass spectrometric identification of sites of interaction for UreD, UreF, and urease.J Biol Chem. 2004 Apr 9;279(15):15305-13. doi: 10.1074/jbc.M312979200. Epub 2004 Jan 28. J Biol Chem. 2004. PMID: 14749331
Cited by
-
Integrated Analysis of Cross-Links and Dead-End Peptides for Enhanced Interpretation of Quantitative XL-MS.J Proteome Res. 2023 Sep 1;22(9):2900-2908. doi: 10.1021/acs.jproteome.3c00191. Epub 2023 Aug 8. J Proteome Res. 2023. PMID: 37552582 Free PMC article.
-
Proteomics-based mass spectrometry profiling of SARS-CoV-2 infection from human nasopharyngeal samples.Mass Spectrom Rev. 2024 Jan-Feb;43(1):193-229. doi: 10.1002/mas.21813. Epub 2022 Sep 29. Mass Spectrom Rev. 2024. PMID: 36177493 Free PMC article. Review.
-
Protein painting for structural and binding site analysis via intracellular lysine reactivity profiling with o-phthalaldehyde.Chem Sci. 2024 Mar 20;15(16):6064-6075. doi: 10.1039/d4sc00032c. eCollection 2024 Apr 24. Chem Sci. 2024. PMID: 38665522 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

