Habitat environmental factors influence intestinal microbial diversity of the short-faced moles (Scaptochirus moschata)
- PMID: 34164757
- PMCID: PMC8222469
- DOI: 10.1186/s13568-021-01252-2
Habitat environmental factors influence intestinal microbial diversity of the short-faced moles (Scaptochirus moschata)
Erratum in
-
Correction to: Habitat environmental factors influence intestinal microbial diversity of the short-faced moles (Scaptochirus moschatus).AMB Express. 2022 Mar 3;12(1):26. doi: 10.1186/s13568-022-01373-2. AMB Express. 2022. PMID: 35239039 Free PMC article. No abstract available.
Abstract
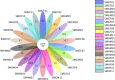
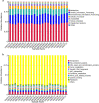
The short-faced moles (Scaptochirus moschata) are unique Chinese mammal that live in burrows for life. They have complex ecological adaptation mechanisms to adapt to perennial underground life. Intestinal microbes play an important role in the ecological adaptation of wild animals. The gut microbiota diversity and its function in short-faced moles' ecological adaptation is a scientific issue worth exploring. In this study, the Illumina HiSeq sequencing platform was used to sequence the V3-V4 hypervariable regions of the 16S rRNA genes of 22 short-faced moles' intestinal samples to study the composition and functional structure of their intestinal microbiota. The results showed that in the short-faced moles' intestine, there are four main phyla, Firmicutes, Proteobacteria, Actinobacteria and Bacteroidete. At the family level, Peptostreptococcaceae and Enterobacteriaceae have the highest abundance. At the genus level, Romboutsia is the genus with the highest microbial abundance. According to the KEGG database, the main functions of short-faced mole gut microbes are metabolism, genetic information processing, environmental information processing, and cellular processes. The function of short-faced mole intestinal microbiota is suitable for its long-term burrowing life. No gender difference is found in the composition and function of the short-faced mole intestinal microbiota. There are significant differences in the composition and functional structure of the short-faced mole gut microbiota between samples collected from different habitats. We conferred that this is related to the different environment factors in which they live, especially to the edaphic factors.
Keywords: Edaphic factor; Gender; Gut microbiome; Sampling locations; Short-faced moles.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. - DOI - PMC - PubMed
-
- Chapman MG, Underwood AJ. Ecological patterns in multivariate assemblages: information and interpretation of negative values in ANOSIM tests. Marine Ecol Progr. 1999;180(3):257–265. doi: 10.3354/meps180257. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

