Evolutionary analysis and functional characterization of SiBRI1 as a Brassinosteroid receptor gene in foxtail millet
- PMID: 34167462
- PMCID: PMC8223282
- DOI: 10.1186/s12870-021-03081-8
Evolutionary analysis and functional characterization of SiBRI1 as a Brassinosteroid receptor gene in foxtail millet
Abstract
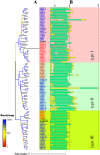
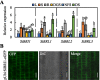
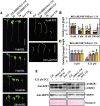
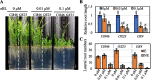
Brassinosteroids (BRs) play important roles in plant growth and development. Although BR receptors have been intensively studied in Arabidopsis, those in foxtail millet remain largely unknown. Here, we show that the BR signaling function of BRASSINOSTEROID INSENSITIVE 1 (BRI1) is conserved between Arabidopsis and foxtail millet, a new model species for C4 and Panicoideae grasses. We identified four putative BR receptor genes in the foxtail millet genome: SiBRI1, SiBRI1-LIKE RECEPTOR KINASE 1 (SiBRL1), SiBRL2 and SiBRL3. Phylogenetic analysis was used to classify the BR receptors in dicots and monocots into three branches. Analysis of their expression patterns by quantitative real-time PCR (qRT-PCR) showed that these receptors were ubiquitously expressed in leaves, stems, dark-grown seedlings, roots and non-flowering spikelets. GFP fusion experiments verified that SiBRI1 localized to the cell membrane. We also explored the SiBRI1 function in Arabidopsis through complementation experiments. Ectopic overexpression of SiBRI1 in an Arabidopsis BR receptor loss-of-function mutant, bri1-116, mostly reversed the developmental defects of the mutant. When SiBRI1 was overexpressed in foxtail millet, the plants showed a drooping leaf phenotype and root development inhibition, lateral root initiation inhibition, and the expression of BR synthesis genes was inhibited. We further identified BRI1-interacting proteins by immunoprecipitation (IP)-mass spectrometry (MS). Our results not only demonstrate that SiBRI1 plays a conserved role in BR signaling in foxtail millet but also provide insight into the molecular mechanism of SiBRI1.
Keywords: BRI1; Brassinosteroids; Foxtail millet; Phylogenetic analysis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Functional Characterization of Soybean Glyma04g39610 as a Brassinosteroid Receptor Gene and Evolutionary Analysis of Soybean Brassinosteroid Receptors.Int J Mol Sci. 2016 Jun 7;17(6):897. doi: 10.3390/ijms17060897. Int J Mol Sci. 2016. PMID: 27338344 Free PMC article.
-
Genome-wide analysis and identification of the low potassium stress responsive gene SiMYB3 in foxtail millet (Setariaitalica L.).BMC Genomics. 2019 Feb 15;20(1):136. doi: 10.1186/s12864-019-5519-2. BMC Genomics. 2019. PMID: 30767761 Free PMC article.
-
DROOPY LEAF1 controls leaf architecture by orchestrating early brassinosteroid signaling.Proc Natl Acad Sci U S A. 2020 Sep 1;117(35):21766-21774. doi: 10.1073/pnas.2002278117. Epub 2020 Aug 17. Proc Natl Acad Sci U S A. 2020. PMID: 32817516 Free PMC article.
-
Ligand perception, activation, and early signaling of plant steroid receptor brassinosteroid insensitive 1.J Integr Plant Biol. 2013 Dec;55(12):1198-211. doi: 10.1111/jipb.12081. Epub 2013 Sep 9. J Integr Plant Biol. 2013. PMID: 23718739 Review.
-
Brassinosteroid signaling and BRI1 dynamics went underground.Curr Opin Plant Biol. 2016 Oct;33:92-100. doi: 10.1016/j.pbi.2016.06.014. Epub 2016 Jul 13. Curr Opin Plant Biol. 2016. PMID: 27419885 Free PMC article. Review.
Cited by
-
Realizing visionary goals for the International Year of Millet (IYoM): accelerating interventions through advances in molecular breeding and multiomics resources.Planta. 2024 Sep 20;260(4):103. doi: 10.1007/s00425-024-04520-0. Planta. 2024. PMID: 39304579 Review.
-
Integrated Lipidomic and Transcriptomic Analysis Reveals Lipid Metabolism in Foxtail Millet (Setaria italica).Front Genet. 2021 Nov 16;12:758003. doi: 10.3389/fgene.2021.758003. eCollection 2021. Front Genet. 2021. PMID: 34868233 Free PMC article.
-
Genetic enhancement of climate-resilient traits in small millets: A review.Heliyon. 2023 Mar 25;9(4):e14502. doi: 10.1016/j.heliyon.2023.e14502. eCollection 2023 Apr. Heliyon. 2023. PMID: 37064482 Free PMC article. Review.
-
Identification of Functional Brassinosteroid Receptor Genes in Oaks and Functional Analysis of QmBRI1.Int J Mol Sci. 2023 Nov 16;24(22):16405. doi: 10.3390/ijms242216405. Int J Mol Sci. 2023. PMID: 38003597 Free PMC article.
-
Multi-omics intervention in Setaria to dissect climate-resilient traits: Progress and prospects.Front Plant Sci. 2022 Aug 31;13:892736. doi: 10.3389/fpls.2022.892736. eCollection 2022. Front Plant Sci. 2022. PMID: 36119586 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

