Targeting the m6A RNA modification pathway blocks SARS-CoV-2 and HCoV-OC43 replication
- PMID: 34168039
- PMCID: PMC8247602
- DOI: 10.1101/gad.348320.121
Targeting the m6A RNA modification pathway blocks SARS-CoV-2 and HCoV-OC43 replication
Abstract
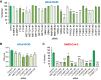
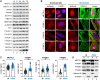
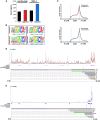
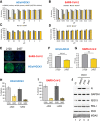
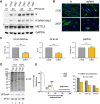
N6-methyladenosine (m6A) is an abundant internal RNA modification, influencing transcript fate and function in uninfected and virus-infected cells. Installation of m6A by the nuclear RNA methyltransferase METTL3 occurs cotranscriptionally; however, the genomes of some cytoplasmic RNA viruses are also m6A-modified. How the cellular m6A modification machinery impacts coronavirus replication, which occurs exclusively in the cytoplasm, is unknown. Here we show that replication of SARS-CoV-2, the agent responsible for the COVID-19 pandemic, and a seasonal human β-coronavirus HCoV-OC43, can be suppressed by depletion of METTL3 or cytoplasmic m6A reader proteins YTHDF1 and YTHDF3 and by a highly specific small molecule METTL3 inhibitor. Reduction of infectious titer correlates with decreased synthesis of viral RNAs and the essential nucleocapsid (N) protein. Sites of m6A modification on genomic and subgenomic RNAs of both viruses were mapped by methylated RNA immunoprecipitation sequencing (meRIP-seq). Levels of host factors involved in m6A installation, removal, and recognition were unchanged by HCoV-OC43 infection; however, nuclear localization of METTL3 and cytoplasmic m6A readers YTHDF1 and YTHDF2 increased. This establishes that coronavirus RNAs are m6A-modified and host m6A pathway components control β-coronavirus replication. Moreover, it illustrates the therapeutic potential of targeting the m6A pathway to restrict coronavirus reproduction.
Keywords: HCoV-OC43; N6-methyladenosine; RNA modification; SARS-CoV-2; coronavirus; direct RNA sequencing; nanopore sequencing; virus–host interactions.
© 2021 Burgess et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
