Thbs1 induces lethal cardiac atrophy through PERK-ATF4 regulated autophagy
- PMID: 34168130
- PMCID: PMC8225674
- DOI: 10.1038/s41467-021-24215-4
Thbs1 induces lethal cardiac atrophy through PERK-ATF4 regulated autophagy
Abstract
The thrombospondin (Thbs) family of secreted matricellular proteins are stress- and injury-induced mediators of cellular attachment dynamics and extracellular matrix protein production. Here we show that Thbs1, but not Thbs2, Thbs3 or Thbs4, induces lethal cardiac atrophy when overexpressed. Mechanistically, Thbs1 binds and activates the endoplasmic reticulum stress effector PERK, inducing its downstream transcription factor ATF4 and causing lethal autophagy-mediated cardiac atrophy. Antithetically, Thbs1-/- mice develop greater cardiac hypertrophy with pressure overload stimulation and show reduced fasting-induced atrophy. Deletion of Thbs1 effectors/receptors, including ATF6α, CD36 or CD47 does not diminish Thbs1-dependent cardiac atrophy. However, deletion of the gene encoding PERK in Thbs1 transgenic mice blunts the induction of ATF4 and autophagy, and largely corrects the lethal cardiac atrophy. Finally, overexpression of PERK or ATF4 using AAV9 gene-transfer similarly promotes cardiac atrophy and lethality. Hence, we identified Thbs1-mediated PERK-eIF2α-ATF4-induced autophagy as a critical regulator of cardiomyocyte size in the stressed heart.
Conflict of interest statement
The authors declare no competing interests.
Figures
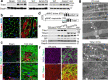






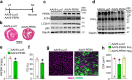

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

